Improving the efficiency of genomic loci capture using oligonucleotide arrays for high throughput resequencing
- PMID: 20043857
- PMCID: PMC2808330
- DOI: 10.1186/1471-2164-10-646
Improving the efficiency of genomic loci capture using oligonucleotide arrays for high throughput resequencing
Abstract
Background: The emergence of next-generation sequencing technology presents tremendous opportunities to accelerate the discovery of rare variants or mutations that underlie human genetic disorders. Although the complete sequencing of the affected individuals' genomes would be the most powerful approach to finding such variants, the cost of such efforts make it impractical for routine use in disease gene research. In cases where candidate genes or loci can be defined by linkage, association, or phenotypic studies, the practical sequencing target can be made much smaller than the whole genome, and it becomes critical to have capture methods that can be used to purify the desired portion of the genome for shotgun short-read sequencing without biasing allelic representation or coverage. One major approach is array-based capture which relies on the ability to create a custom in-situ synthesized oligonucleotide microarray for use as a collection of hybridization capture probes. This approach is being used by our group and others routinely and we are continuing to improve its performance.
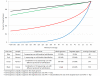
Results: Here, we provide a complete protocol optimized for large aggregate sequence intervals and demonstrate its utility with the capture of all predicted amino acid coding sequence from 3,038 human genes using 241,700 60-mer oligonucleotides. Further, we demonstrate two techniques by which the efficiency of the capture can be increased: by introducing a step to block cross hybridization mediated by common adapter sequences used in sequencing library construction, and by repeating the hybridization capture step. These improvements can boost the targeting efficiency to the point where over 85% of the mapped sequence reads fall within 100 bases of the targeted regions.
Conclusions: The complete protocol introduced in this paper enables researchers to perform practical capture experiments, and includes two novel methods for increasing the targeting efficiency. Coupled with the new massively parallel sequencing technologies, this provides a powerful approach to identifying disease-causing genetic variants that can be localized within the genome by traditional methods.
Figures



References
-
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–380. - PMC - PubMed
-
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. Real-Time DNA Sequencing from Single Polymerase Molecules. Science. 2008;323(5910):133–8. doi: 10.1126/science.1162986. - DOI - PubMed
-
- Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, Causey M, Colonell J, Dimeo J, Efcavitch JW, Giladi E, Gill J, Healy J, Jarosz M, Lapen D, Moulton K, Quake SR, Steinmann K, Thayer E, Tyurina A, Ward R, Weiss H, Xie Z. Single-molecule DNA sequencing of a viral genome. Science. 2008;320(5872):106–109. doi: 10.1126/science.1150427. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

