Frequency of factor H-binding protein modular groups and susceptibility to cross-reactive bactericidal activity in invasive meningococcal isolates
- PMID: 20044056
- PMCID: PMC2830318
- DOI: 10.1016/j.vaccine.2009.12.027
Frequency of factor H-binding protein modular groups and susceptibility to cross-reactive bactericidal activity in invasive meningococcal isolates
Abstract
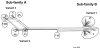
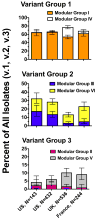
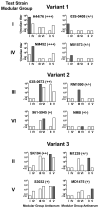
Meningococcal factor H-binding protein (fHbp) is a promising vaccine candidate that elicits serum bactericidal antibodies in humans. Based on sequence variability of the entire protein, fHbp has been divided into three variant groups or two sub-families. We recently reported that the fHbp architecture was modular, consisting of five variable segments, each encoded by genes from one of two lineages. Based on combinations of segments from different lineages, all 70 known fHbp sequence variants could be classified into one of six modular groups. In this study, we analyzed sequences of 172 new fHbp variants that were available from public databases. All but three variants could be classified into one of the six previously described modular groups. Among systematically collected invasive group B isolates from the U.S. and Europe, modular group I overall was most common (60%) but group IV (natural chimeras) accounted for 23% of UK isolates and <1% of U.S. isolates (P<0.0001). Mouse antisera to recombinant fHbp from each of the modular groups showed modular group-specific bactericidal activity against strains with low fHbp expression but had broader activity against strains with higher fHbp expression. Thus both modular group and relative expression of fHbp affected strain susceptibility to anti-fHbp bactericidal activity. The results confirmed the modular architecture of fHbp and underscored its importance for the design of broadly protective group B vaccines in different regions.
Copyright 2009 Elsevier Ltd. All rights reserved.
Figures



References
-
- Tettelin H, Saunders NJ, Heidelberg J, Jeffries AC, Nelson KE, Eisen JA, et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287(5459):1809–15. - PubMed
-
- Grifantini R, Bartolini E, Muzzi A, Draghi M, Frigimelica E, Berger J, et al. Previously unrecognized vaccine candidates against group B meningococcus identified by DNA microarrays. Nat Biotechnol. 2002;20(9):914–21. - PubMed
-
- Hotopp JC, Grifantini R, Kumar N, Tzeng YL, Fouts D, Frigimelica E, et al. Comparative genomics of Neisseria meningitidis: core genome, islands of horizontal transfer and pathogen-specific genes. Microbiology. 2006;152(Pt 12):3733–49. - PubMed
-
- Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287(5459):1816–20. - PubMed
-
- Bernardini G, Braconi D, Lusini P, Santucci A. Postgenomics of Neisseria meningitidis: an update. Expert Rev Proteomics. 2009;6(2):135–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

