PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice
- PMID: 20044539
- PMCID: PMC4295902
- DOI: 10.1126/science.1183439
PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice
Erratum in
- Science. 2010 May 7;328(5979):690
Abstract
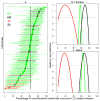
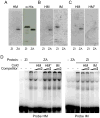
Meiotic recombination events cluster into narrow segments of the genome, defined as hotspots. Here, we demonstrate that a major player for hotspot specification is the Prdm9 gene. First, two mouse strains that differ in hotspot usage are polymorphic for the zinc finger DNA binding array of PRDM9. Second, the human consensus PRDM9 allele is predicted to recognize the 13-mer motif enriched at human hotspots; this DNA binding specificity is verified by in vitro studies. Third, allelic variants of PRDM9 zinc fingers are significantly associated with variability in genome-wide hotspot usage among humans. Our results provide a molecular basis for the distribution of meiotic recombination in mammals, in which the binding of PRDM9 to specific DNA sequences targets the initiation of recombination at specific locations in the genome.
Figures


Comment in
-
Genetics. Genetic control of hotspots.Science. 2010 Feb 12;327(5967):791-2. doi: 10.1126/science.1187155. Science. 2010. PMID: 20150474 No abstract available.
-
PRDM9 points the zinc finger at meiotic recombination hotspots.Genome Biol. 2010;11(2):104. doi: 10.1186/gb-2010-11-2-104. Epub 2010 Feb 26. Genome Biol. 2010. PMID: 20210982 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

