Function of nuclear transport factor 2 and Ran in the 20E signal transduction pathway in the cotton bollworm, Helicoverpa armigera
- PMID: 20044931
- PMCID: PMC2830935
- DOI: 10.1186/1471-2121-11-1
Function of nuclear transport factor 2 and Ran in the 20E signal transduction pathway in the cotton bollworm, Helicoverpa armigera
Abstract
Background: Nuclear transport factor 2 and small GTPase Ran participate in the nucleo-cytoplasm transport of macromolecules, but their function in the 20-hydroxyecdysone (20E) signal transduction pathway are not well known.
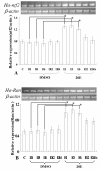
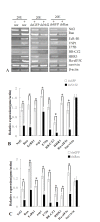
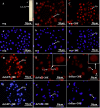
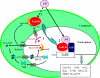
Results: A 703 bp encoding Ntf2 and a 1233 bp encoding Ran full-length cDNAs were cloned from Helicoverpa armigera, and named Ha-Ntf2 and Ha-Ran, respectively. Northern blot and immunoblotting revealed that Ha-Ntf2 had an obviously higher expression levels in the head-thorax and integument of the metamorphically committed larvae. In contrast, the expression of Ha-Ran did not show obvious variation at various developmental stages in four tissues by immunoblotting analysis, except in the midgut, which showed increased expression from 5th-36 h (molting) to 6th-48 h. Both expressions of Ha-Ntf2 and Ha-Ran could be upregulated by 20E in vitro. Immunohistochemistry revealed that Ha-Ntf2 and Ha-Ran were primarily localized in the nucleus of various tissues. Protein binding assay and co-immunoprecipitation indicated that Ha-Ntf2 and Ha-Ran can combine with each other in vitro and in vivo. Knock down of Ha-Ntf2 or Ha-Ran by RNAi resulted in the suppression of other 20E regulated genes including EcR-B1, USP1, E75B, BR-CZ2, HHR3 and Ha-eIF5c. In addition, the knockdown of Ha-Ntf2 resulted in Ha-Ran being prevented in the cytoplasm. The nuclear location of the ecdysone receptor b1 (EcR-B1) was also blocked after the knockdown of Ha-Ntf2 and Ha-Ran.
Conclusion: These evidences suggested that Ha-Ntf2 and Ha-Ran participated in the 20E signal transduction pathway by regulating the location of EcR-B1.
Figures





Similar articles
-
A eukaryotic initiation factor 5C is upregulated during metamorphosis in the cotton bollworm, Helicoverpa armigera.BMC Dev Biol. 2009 Mar 8;9:19. doi: 10.1186/1471-213X-9-19. BMC Dev Biol. 2009. PMID: 19267937 Free PMC article.
-
A new role for nuclear transport factor 2 and Ran: nuclear import of CapG.Traffic. 2008 May;9(5):695-707. doi: 10.1111/j.1600-0854.2008.00720.x. Epub 2008 Feb 7. Traffic. 2008. PMID: 18266911
-
The impacts of classical insect hormones on the expression profiles of a new digestive trypsin-like protease (TLP) from the cotton bollworm, Helicoverpa armigera.Insect Mol Biol. 2009 Aug;18(4):443-52. doi: 10.1111/j.1365-2583.2009.00884.x. Epub 2009 Apr 20. Insect Mol Biol. 2009. PMID: 19469806
-
Phospholipase Cγ1 connects the cell membrane pathway to the nuclear receptor pathway in insect steroid hormone signaling.J Biol Chem. 2014 May 9;289(19):13026-41. doi: 10.1074/jbc.M113.547018. Epub 2014 Apr 1. J Biol Chem. 2014. PMID: 24692553 Free PMC article.
-
Nucleo-cytoplasmic transport of proteins and RNA in plants.Plant Cell Rep. 2011 Feb;30(2):153-76. doi: 10.1007/s00299-010-0928-3. Epub 2010 Oct 20. Plant Cell Rep. 2011. PMID: 20960203 Free PMC article. Review.
Cited by
-
Solexa sequencing based transcriptome analysis of Helicoverpa armigera larvae.Mol Biol Rep. 2012 Dec;39(12):11051-9. doi: 10.1007/s11033-012-2008-y. Epub 2012 Oct 12. Mol Biol Rep. 2012. PMID: 23065207
-
Research Progress on the Regulation of Autophagy and Apoptosis in Insects by Sterol Hormone 20-Hydroxyecdysone.Insects. 2023 Nov 12;14(11):871. doi: 10.3390/insects14110871. Insects. 2023. PMID: 37999070 Free PMC article. Review.
-
Ran Involved in the Development and Reproduction Is a Potential Target for RNA-Interference-Based Pest Management in Nilaparvata lugens.PLoS One. 2015 Nov 10;10(11):e0142142. doi: 10.1371/journal.pone.0142142. eCollection 2015. PLoS One. 2015. PMID: 26554926 Free PMC article.
-
Ras-like family small GTPases genes in Nilaparvata lugens: Identification, phylogenetic analysis, gene expression and function in nymphal development.PLoS One. 2017 Feb 27;12(2):e0172701. doi: 10.1371/journal.pone.0172701. eCollection 2017. PLoS One. 2017. PMID: 28241066 Free PMC article.
-
The nuclear receptor gene E75 plays a key role in regulating the molting process of the spider mite, Tetranychus urticae.Exp Appl Acarol. 2024 Jan;92(1):1-11. doi: 10.1007/s10493-023-00868-2. Epub 2023 Dec 19. Exp Appl Acarol. 2024. PMID: 38112881
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

