Formaldehyde-assisted isolation of regulatory elements
- PMID: 20046543
- PMCID: PMC2800794
- DOI: 10.1002/wsbm.36
Formaldehyde-assisted isolation of regulatory elements
Abstract
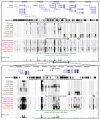
Formaldehyde-Assisted Isolation of Regulatory Elements (FAIRE) is based on locus-specific variations in the ability of protein components of chromatin to trap genomic DNA following formaldehyde treatment. This variation is mostly due to uneven nucleosome distribution since histones are the most abundant and highly crosslinkable components of chromatin. The method can identify and enrich for physically accessible DNA segments of the eukaryotic genome corresponding to known regulatory regions and regions that might have thus far unidentified structural role in the nuclear organization of chromatin. The enrichment patterns are cell type specific and thus might provide information about how transcriptional systems are organized and regulated in various tissues and how they might be disrupted in disease states. Analysis of a 268 kb region of chromosome 19 in human fibroblasts shown here demonstrates that while most DNA fragments detected by FAIRE correspond to sites of DNaseI hypersensitivity in active regions of chromatin, some are found in otherwise repressed chromatin domains and at other sites that are not found with other methods used to probe chromatin structure. Further exploration of FAIRE is warrented due to the simplicity of the protocol and recent advancements in massively parallel sequencing.
Keywords: DNase I sensitivity; FAIRE; chromatin structure; formaldehyde crosslinking; genome organization; nucleosome distribution; regulatory sequences; transcription.
Figures
Similar articles
-
Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements).Methods. 2009 Jul;48(3):233-9. doi: 10.1016/j.ymeth.2009.03.003. Epub 2009 Mar 18. Methods. 2009. PMID: 19303047 Free PMC article.
-
Global Mapping of Open Chromatin Regulatory Elements by Formaldehyde-Assisted Isolation of Regulatory Elements Followed by Sequencing (FAIRE-seq).Methods Mol Biol. 2015;1334:261-72. doi: 10.1007/978-1-4939-2877-4_17. Methods Mol Biol. 2015. PMID: 26404156
-
FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin.Genome Res. 2007 Jun;17(6):877-85. doi: 10.1101/gr.5533506. Epub 2006 Dec 19. Genome Res. 2007. PMID: 17179217 Free PMC article.
-
Formaldehyde crosslinking: a tool for the study of chromatin complexes.J Biol Chem. 2015 Oct 30;290(44):26404-11. doi: 10.1074/jbc.R115.651679. Epub 2015 Sep 9. J Biol Chem. 2015. PMID: 26354429 Free PMC article. Review.
-
Advances of DNase-seq for mapping active gene regulatory elements across the genome in animals.Gene. 2018 Aug 15;667:83-94. doi: 10.1016/j.gene.2018.05.033. Epub 2018 May 14. Gene. 2018. PMID: 29772251 Review.
Cited by
-
Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules.Genome Res. 2012 Dec;22(12):2497-506. doi: 10.1101/gr.143008.112. Epub 2012 Sep 7. Genome Res. 2012. PMID: 22960375 Free PMC article.
-
Maps of open chromatin guide the functional follow-up of genome-wide association signals: application to hematological traits.PLoS Genet. 2011 Jun;7(6):e1002139. doi: 10.1371/journal.pgen.1002139. Epub 2011 Jun 30. PLoS Genet. 2011. PMID: 21738486 Free PMC article.
-
Emerging Approaches to Profile Accessible Chromatin from Formalin-Fixed Paraffin-Embedded Sections.Epigenomes. 2024 May 12;8(2):20. doi: 10.3390/epigenomes8020020. Epigenomes. 2024. PMID: 38804369 Free PMC article. Review.
-
CATA: a comprehensive chromatin accessibility database for cancer.Database (Oxford). 2020 Jan 17;2022:baab085. doi: 10.1093/database/baab085. Database (Oxford). 2020. PMID: 35134148 Free PMC article.
-
Enhancers as information integration hubs in development: lessons from genomics.Trends Genet. 2012 Jun;28(6):276-84. doi: 10.1016/j.tig.2012.02.008. Epub 2012 Apr 7. Trends Genet. 2012. PMID: 22487374 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

