The Electrospray Ionization - Mass Spectra of Erythromycin A Obtained from a Marine Streptomyces sp. Mutant
- PMID: 20046738
- PMCID: PMC2792501
- DOI: 10.4103/0250-474X.42979
The Electrospray Ionization - Mass Spectra of Erythromycin A Obtained from a Marine Streptomyces sp. Mutant
Abstract
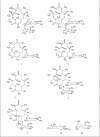
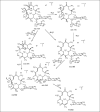
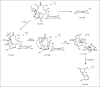
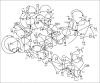
In our ongoing search for production improvements of bioactive secondary metabolites from marine Streptomyces through the induction of mutations using UV light, out of 145 isolates, mutant 10/14 was able to produce potent antibacterial metabolites other than the parent strain as established by chromatographic analysis. Up-scaling fermentation of mutant 10/14, followed by working up and isolation delivered five metabolites, phenazine, 1-acetyl-beta -carboline, perlolyrin and erythromycin A, along with an oily substance. The latter two compounds were responsible for the antibacterial activity of the strain. In this article, we discuss with the mutation of the marine Streptomyces sp. AH2, bioactivity evaluation, fermentation and isolation of the microbial metabolites. Moreover, we study to first time in detail the 1D and 2D NMR and ESI MS data including ESI MS(2) and MS(3) patterns combined with HRESI MS of erythromycin A.
Keywords: ESI-MS/MS; Marine Streptomyces mutation; bioactivity evaluation; erythromycin A.
Figures
Similar articles
-
Metabolite profile of marine-derived endophytic Streptomyces sundarbansensis WR1L1S8 by liquid chromatography-mass spectrometry and evaluation of culture conditions on antibacterial activity and mycelial growth.J Appl Microbiol. 2014 Jan;116(1):39-50. doi: 10.1111/jam.12360. Epub 2013 Nov 11. J Appl Microbiol. 2014. PMID: 24118945
-
Oxoprothracarcin, a novel pyrrolo[1,4]benzodiazepine antibiotic from marine Streptomyces sp. M10946.Drug Discov Ther. 2013 Dec;7(6):243-7. doi: 10.5582/ddt.2013.v7.6.243. Drug Discov Ther. 2013. PMID: 24423655
-
Electrospray ionization mass spectra of piperazimycins A and B and gamma-butyrolactones from a marine-derived Streptomyces sp.J Antibiot (Tokyo). 2008 Dec;61(12):736-46. doi: 10.1038/ja.2008.87. J Antibiot (Tokyo). 2008. PMID: 19194032
-
New nemadectin congener from Streptomyces microflavus neau3: fermentation, isolation, structure elucidation and biological activities.J Antibiot (Tokyo). 2010 Apr;63(4):171-5. doi: 10.1038/ja.2010.12. Epub 2010 Feb 26. J Antibiot (Tokyo). 2010. PMID: 20186170
-
Characterization and mapping of secondary metabolites of Streptomyces sp. from caatinga by desorption electrospray ionization mass spectrometry (DESI-MS).Anal Bioanal Chem. 2018 Nov;410(27):7135-7144. doi: 10.1007/s00216-018-1315-0. Epub 2018 Sep 8. Anal Bioanal Chem. 2018. PMID: 30196421
Cited by
-
A comprehensive review of glycosylated bacterial natural products.Chem Soc Rev. 2015 Nov 7;44(21):7591-697. doi: 10.1039/c4cs00426d. Chem Soc Rev. 2015. PMID: 25735878 Free PMC article. Review.
-
Metabolomic Profiling, In Vitro Antimalarial Investigation and In Silico Modeling of the Marine Actinobacterium Strain Rhodococcus sp. UR111 Associated with the Soft Coral Nephthea sp.Antibiotics (Basel). 2022 Nov 15;11(11):1631. doi: 10.3390/antibiotics11111631. Antibiotics (Basel). 2022. PMID: 36421275 Free PMC article.
References
-
- Laatsch H. In: Marine Bacterial metabolites: Frontiers in marine biotechnology. Proksch P, Müller WEG, editors. Norfolk, UK: Horizon Bioscience; 2006. pp. 225–88.
-
- Mellouli L, Ameur-Mehdi RB, Salem M, Bejar S. Isolation, purification and partial characterization of antibacterial activities produced by a newly isolated Streptomyces sp. US24 strain. Res Microbiol. 2003;154:345–52. - PubMed
-
- Keay BA, Rodrigo R. A convergent synthesis of (±) daunomycinone. Tetrahedron. 1984;40:4597–607.
-
- Sasaki T, Tsukada Y, Deutsch HF, Hirai H. Daunomycin-arachidonic acid complex as a potential new antitumor agent. Cancer Chemother Pharmacol. 1984;13:75–7. - PubMed
-
- Taatjes DJ, Gaudiano G, Resing K, Koch TH. Alkylation of DNA by the anthracycline, antitumor drugs adriamycin and daunomycin. J Med Chem. 1996;39:4135–8. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases