Phosphoproteomics for the masses
- PMID: 20047291
- PMCID: PMC2810156
- DOI: 10.1021/cb900277e
Phosphoproteomics for the masses
Abstract
Protein phosphorylation serves as a primary mechanism of signal transduction in the cells of biological organisms. Technical advancements over the last several years in mass spectrometry now allow for the large-scale identification and quantitation of in vivo phosphorylation at unprecedented levels. These developments have occurred in the areas of sample preparation, instrumentation, quantitative methodology, and informatics so that today, 10 000-20 000 phosphorylation sites can be identified and quantified within a few weeks. With the rapid development and widespread availability of such data, its translation into biological insight and knowledge is a current obstacle. Here we present an overview of how this technology came to be and is currently applied, as well as future challenges for the field.
Figures

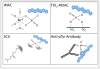
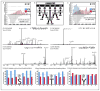

References
-
- Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci. 2002;27:514–520. - PubMed
-
- Moorhead GB, De Wever V, Templeton G, Kerk D. Evolution of protein phosphatases in plants and animals. Biochem J. 2009;417:401–409. - PubMed
-
- Cohen P. The regulation of protein function by multisite phosphorylation--a 25 year update. Trends Biochem Sci. 2000;25:596–601. - PubMed
-
- Sefton BM, Shenolikar S. Overview of protein phosphorylation. Curr Protoc Mol Biol. 2001;Chapter 18(Unit 18):11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

