Roles for cationic residues at the quinolinic acid binding site of quinolinate phosphoribosyltransferase
- PMID: 20047306
- PMCID: PMC2837848
- DOI: 10.1021/bi9018225
Roles for cationic residues at the quinolinic acid binding site of quinolinate phosphoribosyltransferase
Abstract
Quinolinic acid phosphoribosyltransferase (QAPRTase, EC 2.4.2.19) forms nicotinate mononucleotide (NAMN) from quinolinic acid (QA) and 5-phosphoribosyl 1-pyrophosphate (PRPP). Previously determined crystal structures of QAPRTase.QA and QAPRTase.PA.PRPP complexes show positively charged residues (Arg118, Arg152, Arg175, Lys185, and His188) lining the QA binding site. To assess the roles of these residues in the Salmonella typhimurium QAPRTase reaction, they were individually mutated to alanine and the recombinant proteins overexpressed and purified from a recombineered Escherichia coli strain that lacks the QAPRTase gene. Gel filtration indicated that the mutations did not affect the dimeric aggregation state of the enzymes. Arg175 is critical for the QAPRTase reaction, and its mutation to alanine produced an inactive enzyme. The k(cat) values for R152A and K185A were reduced by 33-fold and 625-fold, and binding affinity of PRPP and QA to the enzymes decreased. R152A and K185A mutants displayed 116-fold and 83-fold increases in activity toward the normally inactive QA analogue, nicotinic acid (NA), indicating roles for these residues in defining the substrate specificity of QAPRTase. Moreover, K185A QAPRTase displayed a 300-fold higher k(cat)/K(m) for NA over the natural substrate QA. Pre-steady-state analysis of K185A with QA revealed a burst of nucleotide formation followed by a slower steady-state rate, unlike the linear kinetics of WT. Intriguingly, pre-steady-state analysis of K185A with NA produced a rapid but linear rate for NAMN formation. The result implies a critical role for Lys185 in the chemistry of the QAPRTase intermediate. Arg118 is an essential residue that reaches across the dimer interface. Mutation of Arg118 to alanine resulted in 5000-fold decrease in k(cat) value and a decrease in the binding affinity of QA and PRPP to R152A. Equimolar mixtures of R118A with inactive or virtually inactive mutants produced approximately 50% of the enzymatic activity of WT, establishing an interfacial role for Arg118 during catalysis.
Figures
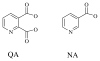






Similar articles
-
Biochemical characterization of quinolinic acid phosphoribosyltransferase from Mycobacterium tuberculosis H37Rv and inhibition of its activity by pyrazinamide.PLoS One. 2014 Jun 20;9(6):e100062. doi: 10.1371/journal.pone.0100062. eCollection 2014. PLoS One. 2014. PMID: 24949952 Free PMC article.
-
Interactions at the 2 and 5 positions of 5-phosphoribosyl pyrophosphate are essential in Salmonella typhimurium quinolinate phosphoribosyltransferase.Biochemistry. 2010 Feb 23;49(7):1377-87. doi: 10.1021/bi9018219. Biochemistry. 2010. PMID: 20047307 Free PMC article.
-
A new function for a common fold: the crystal structure of quinolinic acid phosphoribosyltransferase.Structure. 1997 Jan 15;5(1):47-58. doi: 10.1016/s0969-2126(97)00165-2. Structure. 1997. PMID: 9016724
-
Quinolinate phosphoribosyltransferase: kinetic mechanism for a type II PRTase.Biochemistry. 2002 Mar 12;41(10):3520-8. doi: 10.1021/bi012148g. Biochemistry. 2002. PMID: 11876660
-
Enzymology of NAD+ synthesis.Adv Enzymol Relat Areas Mol Biol. 1999;73:135-82, xi. doi: 10.1002/9780470123195.ch5. Adv Enzymol Relat Areas Mol Biol. 1999. PMID: 10218108 Review.
Cited by
-
Modeling the interaction between quinolinate and the receptor for advanced glycation end products (RAGE): relevance for early neuropathological processes.PLoS One. 2015 Mar 10;10(3):e0120221. doi: 10.1371/journal.pone.0120221. eCollection 2015. PLoS One. 2015. PMID: 25757085 Free PMC article.
-
Biochemical characterization of quinolinic acid phosphoribosyltransferase from Mycobacterium tuberculosis H37Rv and inhibition of its activity by pyrazinamide.PLoS One. 2014 Jun 20;9(6):e100062. doi: 10.1371/journal.pone.0100062. eCollection 2014. PLoS One. 2014. PMID: 24949952 Free PMC article.
References
-
- Musick WD. Structural features of the phosphoribosyltransferases and their relationship to the human deficiency disorders of purine and pyrimidine metabolism. CRC Crit Rev Biochem. 1981;11:1–34. - PubMed
-
- Eads JC, Ozturk D, Wexler TB, Grubmeyer C, Sacchettini JC. A new function for a common fold: the crystal structure of quinolinic acid phosphoribosyltransferase. Structure. 1997;5:47–58. - PubMed
-
- Chappie JS, Canaves JM, Han GW, Rife CL, Xu Q, Stevens RC. The structure of a eukaryotic nicotinic acid phosphoribosyltransferase reveals structural heterogeneity among type II PRTases. Structure. 2005;13:1385–1396. - PubMed
-
- Shin DH, Oganesyan N, Jancarik J, Yokota H, Kim R, Kim SH. Crystal structure of a nicotinate phosphoribosyltransferase from Thermoplasma acidophilum. J Biol Chem. 2005;280:18326–18335. - PubMed
-
- Wang T, Zhang X, Bheda P, Revollo JR, Imai S, Wolberger C. Structure of Nampt/PBEF/visfatin, a mammalian NAD+ biosynthetic enzyme. Nat Struct Mol Biol. 2006;13:661–662. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

