Use of wrapper algorithms coupled with a random forests classifier for variable selection in large-scale genomic association studies
- PMID: 20047492
- PMCID: PMC2980837
- DOI: 10.1089/cmb.2008.0037
Use of wrapper algorithms coupled with a random forests classifier for variable selection in large-scale genomic association studies
Abstract
Modern large-scale genetic association studies generate increasingly high-dimensional datasets. Therefore, some variable selection procedure should be performed before the application of traditional data analysis methods, for reasons of both computational efficiency and problems related to overfitting. We describe here a "wrapper" strategy (SIZEFIT) for variable selection that uses a Random Forests classifier, coupled with various local search/optimization algorithms. We apply it to a large dataset consisting of 2,425 African-American and non-Hispanic white individuals genotyped for 4,869 single-nucleotide polymorphisms (SNPs) in a coronary heart disease (CHD) case-cohort association study (Atherosclerosis Risk in Communities), using incident CHD and plasma low-density lipoprotein (LDL) cholesterol levels as the dependent variables. We show that most SNPs can be safely removed from the dataset without compromising the predictive (classification) accuracy, with only a small number of SNPs (sometimes less than 100) containing any predictive signal. A statistical (SUMSTAT) approach is also applied to the dataset for comparison purposes. We describe a novel method for refining the subset of signal-containing SNPs (FIXFIT), based on an Extremal Optimization algorithm. Finally, we compare the top SNP rankings obtained by different methods and devise practical guidelines for researchers trying to generate a compact subset of predictive SNPs from genome-wide association datasets. Interestingly, there is a significant amount of overlap between seemingly very heterogeneous rankings. We conclude by constructing compact optimal predictive SNP subsets for CHD (less than 150 SNPs) and LDL (less than 300 SNPs) phenotypes, and by comparing various rankings for two well-known positive control SNPs for LDL in the apolipoprotein E gene.
Figures

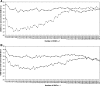

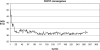

References
-
- ARIC Investigators. The Atherosclerosis Risk in Communities (ARIC) study: design and objectives. Am. J. Epidemiol. 1989;129:687–702. - PubMed
-
- Boettcher S. Percus A.G. Nature's way of optimizing. Artif. Intell. 2000;119:275–286.
-
- Braga-Neto U. Hashimoto R. Dougherty E.R., et al. Is cross-validation better than resubstitution for ranking genes? Bioinformatics. 2004;20:253–258. - PubMed
-
- Breiman L. Random Forests. Mach. Learn. 2001;45:5–32.
Publication types
MeSH terms
Substances
Grants and funding
- U01-HG004402/HG/NHGRI NIH HHS/United States
- N01-HC-55022/HC/NHLBI NIH HHS/United States
- N01-HC-55016/HC/NHLBI NIH HHS/United States
- R37-HL051021/HL/NHLBI NIH HHS/United States
- R01 HL072810/HL/NHLBI NIH HHS/United States
- N01-HC-55020/HC/NHLBI NIH HHS/United States
- R01 HL074735/HL/NHLBI NIH HHS/United States
- P50-GM065509/GM/NIGMS NIH HHS/United States
- R37 HL051021/HL/NHLBI NIH HHS/United States
- U01 HG004402/HG/NHGRI NIH HHS/United States
- N01-HC-55021/HC/NHLBI NIH HHS/United States
- P50 GM065509/GM/NIGMS NIH HHS/United States
- R01-HL072810/HL/NHLBI NIH HHS/United States
- N01-HC-55019/HC/NHLBI NIH HHS/United States
- R01-HL7473501/HL/NHLBI NIH HHS/United States
- N01-HC-55015/HC/NHLBI NIH HHS/United States
- R01-HL87641/HL/NHLBI NIH HHS/United States
- N01-HC-55018/HC/NHLBI NIH HHS/United States
- R01 HL087641/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources

