Amino acid "little Big Bang": representing amino acid substitution matrices as dot products of Euclidian vectors
- PMID: 20047649
- PMCID: PMC3098074
- DOI: 10.1186/1471-2105-11-4
Amino acid "little Big Bang": representing amino acid substitution matrices as dot products of Euclidian vectors
Abstract
Background: Sequence comparisons make use of a one-letter representation for amino acids, the necessary quantitative information being supplied by the substitution matrices. This paper deals with the problem of finding a representation that provides a comprehensive description of amino acid intrinsic properties consistent with the substitution matrices.
Results: We present a Euclidian vector representation of the amino acids, obtained by the singular value decomposition of the substitution matrices. The substitution matrix entries correspond to the dot product of amino acid vectors. We apply this vector encoding to the study of the relative importance of various amino acid physicochemical properties upon the substitution matrices. We also characterize and compare the PAM and BLOSUM series substitution matrices.
Conclusions: This vector encoding introduces a Euclidian metric in the amino acid space, consistent with substitution matrices. Such a numerical description of the amino acid is useful when intrinsic properties of amino acids are necessary, for instance, building sequence profiles or finding consensus sequences, using machine learning algorithms such as Support Vector Machine and Neural Networks algorithms.
Figures

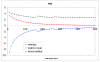
 , obtained as the dot products of the raw, non-centered, vectors. Bottom panel: the blue curve is the same as above but with centered matrix elements (i.e., the mean of the shifted BLOSUM62 matrix is zero), the red curve is the approximation computed with the centered vectors, as described in the text. The x-axis corresponds to the sorted 210 lower triangular matrix elements, e.g., the 210th element is the diagonal element corresponding to the tryptophan, sWW - the largest element in the BLOSUM62 matrix. The y-axis corresponds to the values of the matrix elements. Notice that correlation coefficients are very similar in both cases (0.989 for the curves of the top panel vs 0.998 for the curves of the bottom panel).
, obtained as the dot products of the raw, non-centered, vectors. Bottom panel: the blue curve is the same as above but with centered matrix elements (i.e., the mean of the shifted BLOSUM62 matrix is zero), the red curve is the approximation computed with the centered vectors, as described in the text. The x-axis corresponds to the sorted 210 lower triangular matrix elements, e.g., the 210th element is the diagonal element corresponding to the tryptophan, sWW - the largest element in the BLOSUM62 matrix. The y-axis corresponds to the values of the matrix elements. Notice that correlation coefficients are very similar in both cases (0.989 for the curves of the top panel vs 0.998 for the curves of the bottom panel).
Similar articles
-
The construction of amino acid substitution matrices for the comparison of proteins with non-standard compositions.Bioinformatics. 2005 Apr 1;21(7):902-11. doi: 10.1093/bioinformatics/bti070. Epub 2004 Oct 27. Bioinformatics. 2005. PMID: 15509610
-
The ranging of amino acids substitution matrices of various types in accordance with the alignment accuracy criterion.BMC Bioinformatics. 2020 Sep 14;21(Suppl 11):294. doi: 10.1186/s12859-020-03616-0. BMC Bioinformatics. 2020. PMID: 32921315 Free PMC article.
-
An amino acid substitution matrix for protein conformation identification.J Bioinform Comput Biol. 2006 Jun;4(3):769-82. doi: 10.1142/s0219720006002156. J Bioinform Comput Biol. 2006. PMID: 16960974
-
Substitution scoring matrices for proteins - An overview.Protein Sci. 2020 Nov;29(11):2150-2163. doi: 10.1002/pro.3954. Epub 2020 Oct 12. Protein Sci. 2020. PMID: 32954566 Free PMC article. Review.
-
Amino Acid Encoding Methods for Protein Sequences: A Comprehensive Review and Assessment.IEEE/ACM Trans Comput Biol Bioinform. 2020 Nov-Dec;17(6):1918-1931. doi: 10.1109/TCBB.2019.2911677. Epub 2020 Dec 8. IEEE/ACM Trans Comput Biol Bioinform. 2020. PMID: 30998480 Review.
Cited by
-
Insight into neutral and disease-associated human genetic variants through interpretable predictors.PLoS One. 2015 Mar 31;10(3):e0120729. doi: 10.1371/journal.pone.0120729. eCollection 2015. PLoS One. 2015. PMID: 25826299 Free PMC article.
-
How Fitch-Margoliash Algorithm can Benefit from Multi Dimensional Scaling.Evol Bioinform Online. 2011;7:61-85. doi: 10.4137/EBO.S7048. Epub 2011 Jun 7. Evol Bioinform Online. 2011. PMID: 21697992 Free PMC article.
-
Amino acid properties conserved in molecular evolution.PLoS One. 2014 Jun 26;9(6):e98983. doi: 10.1371/journal.pone.0098983. eCollection 2014. PLoS One. 2014. PMID: 24967708 Free PMC article.
-
The Ancient Operational Code is Embedded in the Amino Acid Substitution Matrix and aaRS Phylogenies.J Mol Evol. 2020 Mar;88(2):136-150. doi: 10.1007/s00239-019-09918-z. Epub 2019 Nov 28. J Mol Evol. 2020. PMID: 31781936
References
-
- Dayhoff M, Schwartz R, Orcutt B. In: Atlas of protein sequence and structure. Dayhoff M, editor. Vol. 5. National Biomedical Research Fundation, Washington, DC; 1978. A model of evolutionary change in proteins; pp. 345–352.
-
- Maetschke S, Towsey M, Boden M. BLOMAP: an Encoding of Amino Acids which improves Signal Peptide Cleavage Site Prediction. Asia Pacific Bioinformatics Conference. 2005. pp. 141–150. full_text.
-
- Swanson R. A vector representation for amino acid sequences. Bull Math Biol. 1984;46:623–639. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

