Amino acid "little Big Bang": representing amino acid substitution matrices as dot products of Euclidian vectors
- PMID: 20047649
- PMCID: PMC3098074
- DOI: 10.1186/1471-2105-11-4
Amino acid "little Big Bang": representing amino acid substitution matrices as dot products of Euclidian vectors
Abstract
Background: Sequence comparisons make use of a one-letter representation for amino acids, the necessary quantitative information being supplied by the substitution matrices. This paper deals with the problem of finding a representation that provides a comprehensive description of amino acid intrinsic properties consistent with the substitution matrices.
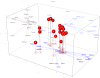
Results: We present a Euclidian vector representation of the amino acids, obtained by the singular value decomposition of the substitution matrices. The substitution matrix entries correspond to the dot product of amino acid vectors. We apply this vector encoding to the study of the relative importance of various amino acid physicochemical properties upon the substitution matrices. We also characterize and compare the PAM and BLOSUM series substitution matrices.
Conclusions: This vector encoding introduces a Euclidian metric in the amino acid space, consistent with substitution matrices. Such a numerical description of the amino acid is useful when intrinsic properties of amino acids are necessary, for instance, building sequence profiles or finding consensus sequences, using machine learning algorithms such as Support Vector Machine and Neural Networks algorithms.
Figures

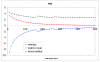
 , obtained as the dot products of the raw, non-centered, vectors. Bottom panel: the blue curve is the same as above but with centered matrix elements (i.e., the mean of the shifted BLOSUM62 matrix is zero), the red curve is the approximation computed with the centered vectors, as described in the text. The x-axis corresponds to the sorted 210 lower triangular matrix elements, e.g., the 210th element is the diagonal element corresponding to the tryptophan, sWW - the largest element in the BLOSUM62 matrix. The y-axis corresponds to the values of the matrix elements. Notice that correlation coefficients are very similar in both cases (0.989 for the curves of the top panel vs 0.998 for the curves of the bottom panel).
, obtained as the dot products of the raw, non-centered, vectors. Bottom panel: the blue curve is the same as above but with centered matrix elements (i.e., the mean of the shifted BLOSUM62 matrix is zero), the red curve is the approximation computed with the centered vectors, as described in the text. The x-axis corresponds to the sorted 210 lower triangular matrix elements, e.g., the 210th element is the diagonal element corresponding to the tryptophan, sWW - the largest element in the BLOSUM62 matrix. The y-axis corresponds to the values of the matrix elements. Notice that correlation coefficients are very similar in both cases (0.989 for the curves of the top panel vs 0.998 for the curves of the bottom panel).
References
-
- Dayhoff M, Schwartz R, Orcutt B. In: Atlas of protein sequence and structure. Dayhoff M, editor. Vol. 5. National Biomedical Research Fundation, Washington, DC; 1978. A model of evolutionary change in proteins; pp. 345–352.
-
- Maetschke S, Towsey M, Boden M. BLOMAP: an Encoding of Amino Acids which improves Signal Peptide Cleavage Site Prediction. Asia Pacific Bioinformatics Conference. 2005. pp. 141–150. full_text.
-
- Swanson R. A vector representation for amino acid sequences. Bull Math Biol. 1984;46:623–639. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

