The BridgeDb framework: standardized access to gene, protein and metabolite identifier mapping services
- PMID: 20047655
- PMCID: PMC2824678
- DOI: 10.1186/1471-2105-11-5
The BridgeDb framework: standardized access to gene, protein and metabolite identifier mapping services
Abstract
Background: Many complementary solutions are available for the identifier mapping problem. This creates an opportunity for bioinformatics tool developers. Tools can be made to flexibly support multiple mapping services or mapping services could be combined to get broader coverage. This approach requires an interface layer between tools and mapping services.
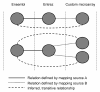
Results: Here we present BridgeDb, a software framework for gene, protein and metabolite identifier mapping. This framework provides a standardized interface layer through which bioinformatics tools can be connected to different identifier mapping services. This approach makes it easier for tool developers to support identifier mapping. Mapping services can be combined or merged to support multi-omics experiments or to integrate custom microarray annotations. BridgeDb provides its own ready-to-go mapping services, both in webservice and local database forms. However, the framework is intended for customization and adaptation to any identifier mapping service. BridgeDb has already been integrated into several bioinformatics applications.
Conclusion: By uncoupling bioinformatics tools from mapping services, BridgeDb improves capability and flexibility of those tools. All described software is open source and available at http://www.bridgedb.org.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

