A markov classification model for metabolic pathways
- PMID: 20047667
- PMCID: PMC2823754
- DOI: 10.1186/1748-7188-5-10
A markov classification model for metabolic pathways
Abstract
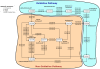
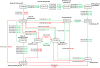
Background: This paper considers the problem of identifying pathways through metabolic networks that relate to a specific biological response. Our proposed model, HME3M, first identifies frequently traversed network paths using a Markov mixture model. Then by employing a hierarchical mixture of experts, separate classifiers are built using information specific to each path and combined into an ensemble prediction for the response.
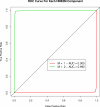
Results: We compared the performance of HME3M with logistic regression and support vector machines (SVM) for both simulated pathways and on two metabolic networks, glycolysis and the pentose phosphate pathway for Arabidopsis thaliana. We use AltGenExpress microarray data and focus on the pathway differences in the developmental stages and stress responses of Arabidopsis. The results clearly show that HME3M outperformed the comparison methods in the presence of increasing network complexity and pathway noise. Furthermore an analysis of the paths identified by HME3M for each metabolic network confirmed known biological responses of Arabidopsis.
Conclusions: This paper clearly shows HME3M to be an accurate and robust method for classifying metabolic pathways. HME3M is shown to outperform all comparison methods and further is capable of identifying known biologically active pathways within microarray data.
Figures







Similar articles
-
Mining metabolic pathways through gene expression.Bioinformatics. 2010 Sep 1;26(17):2128-35. doi: 10.1093/bioinformatics/btq344. Epub 2010 Jun 29. Bioinformatics. 2010. PMID: 20587705 Free PMC article.
-
Active pathway identification and classification with probabilistic ensembles.Genome Inform. 2010 Jan;22:30-40. Genome Inform. 2010. PMID: 20238417
-
A hierarchical mixture of Markov models for finding biologically active metabolic paths using gene expression and protein classes.Proc IEEE Comput Syst Bioinform Conf. 2004:341-52. doi: 10.1109/csb.2004.1332447. Proc IEEE Comput Syst Bioinform Conf. 2004. PMID: 16448027
-
Identifying pathways of coordinated gene expression.Methods Mol Biol. 2013;939:69-85. doi: 10.1007/978-1-62703-107-3_7. Methods Mol Biol. 2013. PMID: 23192542
-
Reviewing ensemble classification methods in breast cancer.Comput Methods Programs Biomed. 2019 Aug;177:89-112. doi: 10.1016/j.cmpb.2019.05.019. Epub 2019 May 20. Comput Methods Programs Biomed. 2019. PMID: 31319964 Review.
Cited by
-
Mining metabolic pathways through gene expression.Bioinformatics. 2010 Sep 1;26(17):2128-35. doi: 10.1093/bioinformatics/btq344. Epub 2010 Jun 29. Bioinformatics. 2010. PMID: 20587705 Free PMC article.
References
-
- Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, Holloway E, Kapushesky M, Kemmeren P, Lara GG, Oezcimen A, Rocca-Serra P, Sansone S. ArrayExpress-a public repository for microarray gene expression data at the EBI. Nucl Acids Res. 2003;31:68–71. doi: 10.1093/nar/gkg091. - DOI - PMC - PubMed
-
- Pireddu L, Poulin B, Szafron D, Lu P, Wishart DS. Pathway Analyst - Automated Metabolic Pathway Prediction. Proceedings of the 2005 IEEE Symposium on Computational Intelligence. 2005. http://metabolomics.ca/News/publications/2005cibcb-path.pdf
-
- Jordan M. Learning in Graphical Models. Norwell, MD: Kluwer Academic Publishers; 1998.
LinkOut - more resources
Full Text Sources

