Genomic characterization of the Yersinia genus
- PMID: 20047673
- PMCID: PMC2847712
- DOI: 10.1186/gb-2010-11-1-r1
Genomic characterization of the Yersinia genus
Abstract
Background: New DNA sequencing technologies have enabled detailed comparative genomic analyses of entire genera of bacterial pathogens. Prior to this study, three species of the enterobacterial genus Yersinia that cause invasive human diseases (Yersinia pestis, Yersinia pseudotuberculosis, and Yersinia enterocolitica) had been sequenced. However, there were no genomic data on the Yersinia species with more limited virulence potential, frequently found in soil and water environments.
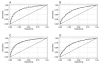
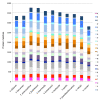
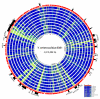
Results: We used high-throughput sequencing-by-synthesis instruments to obtain 25- to 42-fold average redundancy, whole-genome shotgun data from the type strains of eight species: Y. aldovae, Y. bercovieri, Y. frederiksenii, Y. kristensenii, Y. intermedia, Y. mollaretii, Y. rohdei, and Y. ruckeri. The deepest branching species in the genus, Y. ruckeri, causative agent of red mouth disease in fish, has the smallest genome (3.7 Mb), although it shares the same core set of approximately 2,500 genes as the other members of the species, whose genomes range in size from 4.3 to 4.8 Mb. Yersinia genomes had a similar global partition of protein functions, as measured by the distribution of Cluster of Orthologous Groups families. Genome to genome variation in islands with genes encoding functions such as ureases, hydrogenases and B-12 cofactor metabolite reactions may reflect adaptations to colonizing specific host habitats.
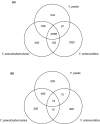
Conclusions: Rapid high-quality draft sequencing was used successfully to compare pathogenic and non-pathogenic members of the Yersinia genus. This work underscores the importance of the acquisition of horizontally transferred genes in the evolution of Y. pestis and points to virulence determinants that have been gained and lost on multiple occasions in the history of the genus.
Figures
References
-
- Ecker DJ, Sampath R, Willett P, Wyatt JR, Samant V, Massire C, Hall TA, Hari K, McNeil JA, Buchen-Osmond C, Budowle B. The Microbial Rosetta Stone Database: a compilation of global and emerging infectious microorganisms and bioterrorist threat agents. BMC Microbiol. 2005;5:19. doi: 10.1186/1471-2180-5-19. - DOI - PMC - PubMed
-
- Van Ert MN, Easterday WR, Huynh LY, Okinaka RT, Hugh-Jones ME, Ravel J, Zanecki SR, Pearson T, Simonson TS, U'Ren JM, Kachur SM, Leadem-Dougherty RR, Rhoton SD, Zinser G, Farlow J, Coker PR, Smith KL, Wang B, Kenefic LJ, Fraser-Liggett CM, Wagner DM, Keim P. Global Genetic Population Structure of Bacillus anthracis. PLoS ONE. 2007;2:e461. doi: 10.1371/journal.pone.0000461. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

