Complete genome sequence of the fire blight pathogen Erwinia pyrifoliae DSM 12163T and comparative genomic insights into plant pathogenicity
- PMID: 20047678
- PMCID: PMC2827408
- DOI: 10.1186/1471-2164-11-2
Complete genome sequence of the fire blight pathogen Erwinia pyrifoliae DSM 12163T and comparative genomic insights into plant pathogenicity
Abstract
Background: Erwinia pyrifoliae is a newly described necrotrophic pathogen, which causes fire blight on Asian (Nashi) pear and is geographically restricted to Eastern Asia. Relatively little is known about its genetics compared to the closely related main fire blight pathogen E. amylovora.
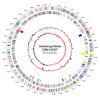
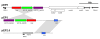
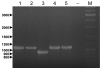
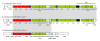
Results: The genome of the type strain of E. pyrifoliae strain DSM 12163T, was sequenced using both 454 and Solexa pyrosequencing and annotated. The genome contains a circular chromosome of 4.026 Mb and four small plasmids. Based on their respective role in virulence in E. amylovora or related organisms, we identified several putative virulence factors, including type III and type VI secretion systems and their effectors, flagellar genes, sorbitol metabolism, iron uptake determinants, and quorum-sensing components. A deletion in the rpoS gene covering the most conserved region of the protein was identified which may contribute to the difference in virulence/host-range compared to E. amylovora. Comparative genomics with the pome fruit epiphyte Erwinia tasmaniensis Et1/99 showed that both species are overall highly similar, although specific differences were identified, for example the presence of some phage gene-containing regions and a high number of putative genomic islands containing transposases in the E. pyrifoliae DSM 12163T genome.
Conclusions: The E. pyrifoliae genome is an important addition to the published genome of E. tasmaniensis and the unfinished genome of E. amylovora providing a foundation for re-sequencing additional strains that may shed light on the evolution of the host-range and virulence/pathogenicity of this important group of plant-associated bacteria.
Figures



References
-
- Duffy B, Schärer H-J, Bünter M, Klay A, Holliger E. Regulatory measures against Erwinia amylovora in Switzerland. EPPO Bull. 2005;35:239–244. doi: 10.1111/j.1365-2338.2005.00820.x. - DOI
-
- Bonn WG, Zwet T van der. In: Fire blight the disease and its causative agent, Erwinia amylovora. Vanneste JL, editor. Wallingford, UK CAB International; 2000. Distribution and economic importance of fire blight; pp. 37–53. full_text.
-
- Thomson SV. In: Fire blight the disease and its causative agent, Erwinia amylovora. Vanneste JL, editor. Wallingford, UK CAB International; 2000. Epidemiology of fire blight; pp. 9–36. full_text.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

