Analysis of newly established EST databases reveals similarities between heart regeneration in newt and fish
- PMID: 20047682
- PMCID: PMC2823690
- DOI: 10.1186/1471-2164-11-4
Analysis of newly established EST databases reveals similarities between heart regeneration in newt and fish
Abstract
Background: The newt Notophthalmus viridescens possesses the remarkable ability to respond to cardiac damage by formation of new myocardial tissue. Surprisingly little is known about changes in gene activities that occur during the course of regeneration. To begin to decipher the molecular processes, that underlie restoration of functional cardiac tissue, we generated an EST database from regenerating newt hearts and compared the transcriptional profile of selected candidates with genes deregulated during zebrafish heart regeneration.
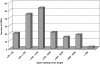
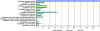
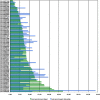
Results: A cDNA library of 100,000 cDNA clones was generated from newt hearts 14 days after ventricular injury. Sequencing of 11520 cDNA clones resulted in 2894 assembled contigs. BLAST searches revealed 1695 sequences with potential homology to sequences from the NCBI database. BLAST searches to TrEMBL and Swiss-Prot databases assigned 1116 proteins to Gene Ontology terms. We also identified a relatively large set of 174 ORFs, which are likely to be unique for urodele amphibians. Expression analysis of newt-zebrafish homologues confirmed the deregulation of selected genes during heart regeneration. Sequences, BLAST results and GO annotations were visualized in a relational web based database followed by grouping of identified proteins into clusters of GO Terms. Comparison of data from regenerating zebrafish hearts identified biological processes, which were uniformly overrepresented during cardiac regeneration in newt and zebrafish.
Conclusion: We concluded that heart regeneration in newts and zebrafish led to the activation of similar sets of genes, which suggests that heart regeneration in both species might follow similar principles. The design of the newly established newt EST database allows identification of molecular pathways important for heart regeneration.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

