Normal colon epithelium: a dataset for the analysis of gene expression and alternative splicing events in colon disease
- PMID: 20047688
- PMCID: PMC2823691
- DOI: 10.1186/1471-2164-11-5
Normal colon epithelium: a dataset for the analysis of gene expression and alternative splicing events in colon disease
Abstract
Background: Studies using microarray analysis of colorectal cancer have been generally beleaguered by the lack of a normal cell population of the same lineage as the tumor cell. One of the main objectives of this study was to generate a reference gene expression data set for normal colonic epithelium which can be used in comparisons with diseased tissues, as well as to provide a dataset that could be used as a baseline for studies in alternative splicing.
Results: We present a dependable expression reference data set for non-neoplastic colonic epithelial cells. An enriched population of fresh colon epithelial cells were obtained from non-neoplastic, colectomy specimens and analyzed using Affymetrix GeneChip EXON 1.0 ST arrays. For demonstration purposes, we have compared the data derived from these cells to a publically available set of tumor and matched normal colon data. This analysis allowed an assessment of global gene expression alterations and demonstrated that adjacent normal tissues, with a high degree of cellular heterogeneity, are not always representative of normal cells for comparison to tumors which arise from the colon epithelium. We also examined alternative splicing events in tumors compared to normal colon epithelial cells.
Conclusions: The findings from this study represent the first comprehensive expression profile for non-neoplastic colonic epithelial cells reported. Our analysis of splice variants illustrate that this is a very labor intensive procedure, requiring vigilant examination of the data. It is projected that the contribution of this set of data derived from pure colonic epithelial cells will enhance studies in colon-related disease and offer a vital baseline for studies aimed at elucidating the mechanisms of alternative splicing.
Figures


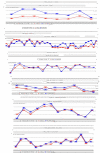
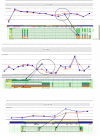
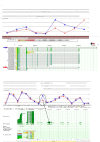

Similar articles
-
An integrative framework identifies alternative splicing events in colorectal cancer development.Mol Oncol. 2014 Feb;8(1):129-41. doi: 10.1016/j.molonc.2013.10.004. Epub 2013 Oct 19. Mol Oncol. 2014. PMID: 24189147 Free PMC article.
-
Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array.BMC Genomics. 2006 Dec 27;7:325. doi: 10.1186/1471-2164-7-325. BMC Genomics. 2006. PMID: 17192196 Free PMC article.
-
Alternative splicing in colon, bladder, and prostate cancer identified by exon array analysis.Mol Cell Proteomics. 2008 Jul;7(7):1214-24. doi: 10.1074/mcp.M700590-MCP200. Epub 2008 Mar 18. Mol Cell Proteomics. 2008. PMID: 18353764
-
A statistical method for predicting splice variants between two groups of samples using GeneChip expression array data.Theor Biol Med Model. 2006 Apr 7;3:19. doi: 10.1186/1742-4682-3-19. Theor Biol Med Model. 2006. PMID: 16603076 Free PMC article.
-
Development, validation and implementation of an in vitro model for the study of metabolic and immune function in normal and inflamed human colonic epithelium.Dan Med J. 2015 Jan;62(1):B4973. Dan Med J. 2015. PMID: 25557335 Review.
Cited by
-
Profiles of alternative splicing in colorectal cancer and their clinical significance: A study based on large-scale sequencing data.EBioMedicine. 2018 Oct;36:183-195. doi: 10.1016/j.ebiom.2018.09.021. Epub 2018 Sep 19. EBioMedicine. 2018. PMID: 30243491 Free PMC article.
-
Colonic epithelial cell diversity in health and inflammatory bowel disease.Nature. 2019 Mar;567(7746):49-55. doi: 10.1038/s41586-019-0992-y. Epub 2019 Feb 27. Nature. 2019. PMID: 30814735
-
Membrane recruitment of the polarity protein Scribble by the cell adhesion receptor TMIGD1.Commun Biol. 2023 Jul 10;6(1):702. doi: 10.1038/s42003-023-05088-3. Commun Biol. 2023. PMID: 37430142 Free PMC article.
-
An integrative framework identifies alternative splicing events in colorectal cancer development.Mol Oncol. 2014 Feb;8(1):129-41. doi: 10.1016/j.molonc.2013.10.004. Epub 2013 Oct 19. Mol Oncol. 2014. PMID: 24189147 Free PMC article.
-
Aberrant gene expression in mucosa adjacent to tumor reveals a molecular crosstalk in colon cancer.Mol Cancer. 2014 Mar 5;13:46. doi: 10.1186/1476-4598-13-46. Mol Cancer. 2014. PMID: 24597571 Free PMC article.
References
-
- Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional Gene Expression Profiles of Colorectal Adenoma, Adenocarcinoma, and Normal Tissue Examined by Oligonucleotide Arrays. Cancer Res. 2001;61(7):3124–30. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

