A duplicate gene rooting of seed plants and the phylogenetic position of flowering plants
- PMID: 20047866
- PMCID: PMC2838261
- DOI: 10.1098/rstb.2009.0233
A duplicate gene rooting of seed plants and the phylogenetic position of flowering plants
Abstract
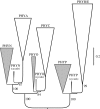
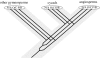
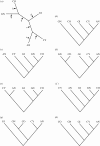
Flowering plants represent the most significant branch in the tree of land plants, with respect to the number of extant species, their impact on the shaping of modern ecosystems and their economic importance. However, unlike so many persistent phylogenetic problems that have yielded to insights from DNA sequence data, the mystery surrounding the origin of angiosperms has deepened with the advent and advance of molecular systematics. Strong statistical support for competing hypotheses and recent novel trees from molecular data suggest that the accuracy of current molecular trees requires further testing. Analyses of phytochrome amino acids using a duplicate gene-rooting approach yield trees that unite cycads and angiosperms in a clade that is sister to a clade in which Gingko and Cupressophyta are successive sister taxa to gnetophytes plus Pinaceae. Application of a cycads + angiosperms backbone constraint in analyses of a morphological dataset yields better resolved trees than do analyses in which extant gymnosperms are forced to be monophyletic. The results have implications both for our assessment of uncertainty in trees from sequence data and for our use of molecular constraints as a way to integrate insights from morphological and molecular evidence.
Figures


Similar articles
-
Single-Copy Genes as Molecular Markers for Phylogenomic Studies in Seed Plants.Genome Biol Evol. 2017 May 1;9(5):1130-1147. doi: 10.1093/gbe/evx070. Genome Biol Evol. 2017. PMID: 28460034 Free PMC article.
-
Phylogeny of seed plants based on all three genomic compartments: extant gymnosperms are monophyletic and Gnetales' closest relatives are conifers.Proc Natl Acad Sci U S A. 2000 Apr 11;97(8):4092-7. doi: 10.1073/pnas.97.8.4092. Proc Natl Acad Sci U S A. 2000. PMID: 10760278 Free PMC article.
-
Phylogenomics resolves the deep phylogeny of seed plants and indicates partial convergent or homoplastic evolution between Gnetales and angiosperms.Proc Biol Sci. 2018 Jun 27;285(1881):20181012. doi: 10.1098/rspb.2018.1012. Proc Biol Sci. 2018. PMID: 29925623 Free PMC article.
-
Perspective: the origin of flowering plants and their reproductive biology--a tale of two phylogenies.Evolution. 2001 Feb;55(2):217-31. doi: 10.1111/j.0014-3820.2001.tb01288.x. Evolution. 2001. PMID: 11308081 Review.
-
Missing links: the genetic architecture of flowers [correction of flower] and floral diversification.Trends Plant Sci. 2002 Jan;7(1):22-31; dicussion 31-4. doi: 10.1016/s1360-1385(01)02098-2. Trends Plant Sci. 2002. PMID: 11804823 Review.
Cited by
-
Light- and hormone-mediated development in non-flowering plants: An overview.Planta. 2020 Nov 27;253(1):1. doi: 10.1007/s00425-020-03501-3. Planta. 2020. PMID: 33245411 Review.
-
Ginkgo biloba's footprint of dynamic Pleistocene history dates back only 390,000 years ago.BMC Genomics. 2018 Apr 27;19(1):299. doi: 10.1186/s12864-018-4673-2. BMC Genomics. 2018. PMID: 29703145 Free PMC article.
-
Incongruence in the phylogenomics era.Nat Rev Genet. 2023 Dec;24(12):834-850. doi: 10.1038/s41576-023-00620-x. Epub 2023 Jun 27. Nat Rev Genet. 2023. PMID: 37369847 Free PMC article. Review.
-
Uncovering dynamic evolution in the plastid genome of seven Ligusticum species provides insights into species discrimination and phylogenetic implications.Sci Rep. 2021 Jan 13;11(1):988. doi: 10.1038/s41598-020-80225-0. Sci Rep. 2021. PMID: 33441833 Free PMC article.
-
Multiple measures could alleviate long-branch attraction in phylogenomic reconstruction of Cupressoideae (Cupressaceae).Sci Rep. 2017 Jan 25;7:41005. doi: 10.1038/srep41005. Sci Rep. 2017. PMID: 28120880 Free PMC article.
References
-
- Abascal F., Posada D., Zardoya R.2007MtArt: a new model of amino acid replacement for Arthropoda. Mol. Biol. Evol 24, 1 (doi:10.1093/molbev/msl136) - DOI - PubMed
-
- Bailey I. W.1944The development of vessels in angiosperms and its significance in morphological research. Am. J. Bot. 31, 421–428 (doi:10.2307/2437302) - DOI
-
- Baldauf S. L., Palmer J. D., Doolittle W. F.1996The root of the universal tree and the origin of eukaryotes based on elongation factor phylogeny. Proc. Natl Acad. Sci. USA 93, 7749–7754 (doi:10.1073/pnas.93.15.7749) - DOI - PMC - PubMed
-
- Barkman T. J., Chenery G., McNeal J. R., Lyons-Weiler J., Ellisens W. J., Moore G., Wolfe A. D., dePamphilis C. W.2000Independent and combined analyses of sequences from all three genomic compartments converge on the root of flowering plant phylogeny. Proc. Natl Acad. Sci. USA 97, 13 166–13 171 (doi:10.1073/pnas.220427497) - DOI - PMC - PubMed
-
- Bierhorst D. W.1971Morphology of vascular plants New York, NY: Macmillan
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

