SnAvi--a new tandem tag for high-affinity protein-complex purification
- PMID: 20047968
- PMCID: PMC2847239
- DOI: 10.1093/nar/gkp1178
SnAvi--a new tandem tag for high-affinity protein-complex purification
Abstract
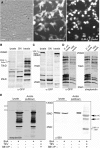
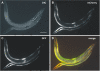
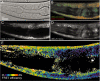
Systematic tandem-affinity-purification (TAP) of protein complexes was tremendously successful in yeast and has changed the general concept of how we understand protein function in eukaryotic cells. The transfer of this method to other model organisms has been difficult and may require specific adaptations. We were especially interested to establish a cell-type-specific TAP system for Caenorhabditis elegans, a model animal well suited to high-throughput analysis, proteomics and systems biology. By combining the high-affinity interaction between in vivo biotinylated target-proteins and streptavidin with the usage of a newly identified epitope of the publicly shared SB1 monoclonal antibody we created a novel in vivo fluorescent tag, the SnAvi-Tag. We show the versatile application of the SnAvi-Tag in Escherichia coli, vertebrate cells and in C. elegans for tandem affinity purification of protein complexes, western blotting and also for the in vivo sub-cellular localization of labelled proteins.
Figures


Similar articles
-
A combined approach for the localization and tandem affinity purification of protein complexes from metazoans.Sci STKE. 2005 Jan 11;2005(266):pl1. doi: 10.1126/stke.2662005pl1. Sci STKE. 2005. PMID: 15644491
-
Targeted purification of SnAvi-tagged proteins.Methods Mol Biol. 2014;1177:163-74. doi: 10.1007/978-1-4939-1034-2_13. Methods Mol Biol. 2014. PMID: 24943322
-
Tandem affinity purification of protein complexes from mammalian cells by the Strep/FLAG (SF)-TAP tag.Methods Mol Biol. 2009;564:359-72. doi: 10.1007/978-1-60761-157-8_21. Methods Mol Biol. 2009. PMID: 19544034
-
Interaction proteomics: characterization of protein complexes using tandem affinity purification-mass spectrometry.Biochem Soc Trans. 2010 Aug;38(4):883-7. doi: 10.1042/BST0380883. Biochem Soc Trans. 2010. PMID: 20658971 Review.
-
Affinity Tags for Protein Purification.Curr Protein Pept Sci. 2020;21(8):821-830. doi: 10.2174/1389203721666200606220109. Curr Protein Pept Sci. 2020. PMID: 32504500 Review.
Cited by
-
Interactome analysis of Caenorhabditis elegans synapses by TurboID-based proximity labeling.J Biol Chem. 2021 Sep;297(3):101094. doi: 10.1016/j.jbc.2021.101094. Epub 2021 Aug 18. J Biol Chem. 2021. PMID: 34416233 Free PMC article.
-
Effective targeting of microglial P2X7 following intracerebroventricular delivery of nanobodies and nanobody-encoding AAVs.Front Pharmacol. 2022 Oct 10;13:1029236. doi: 10.3389/fphar.2022.1029236. eCollection 2022. Front Pharmacol. 2022. PMID: 36299894 Free PMC article.
-
Using C. elegans to identify the protease targets of serpins in vivo.Methods Enzymol. 2011;499:283-99. doi: 10.1016/B978-0-12-386471-0.00014-6. Methods Enzymol. 2011. PMID: 21683259 Free PMC article.
-
Proteomic profiling of cardiac tissue by isolation of nuclei tagged in specific cell types (INTACT).Development. 2014 Feb;141(4):962-73. doi: 10.1242/dev.098327. Development. 2014. PMID: 24496632 Free PMC article.
-
Isolation of serpin-interacting proteins in C. elegans using protein affinity purification.Methods. 2014 Aug 1;68(3):536-41. doi: 10.1016/j.ymeth.2014.04.019. Epub 2014 May 2. Methods. 2014. PMID: 24798811 Free PMC article.
References
-
- Polanowska J, Martin JS, Fisher R, Scopa T, Rae I, Boulton SJ. Tandem immunoaffinity purification of protein complexes from Caenorhabditis elegans. Biotechniques. 2004;36:778–780, 782. - PubMed
-
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. - PubMed
-
- Cheeseman IM, Desai A. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE. 2005;2005:pl1. - PubMed
-
- Terpe K. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2003;60:523–533. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

