C/EBPbetaDeltauORF mice--a genetic model for uORF-mediated translational control in mammals
- PMID: 20047998
- PMCID: PMC2802187
- DOI: 10.1101/gad.557910
C/EBPbetaDeltauORF mice--a genetic model for uORF-mediated translational control in mammals
Abstract
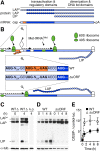
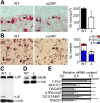
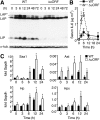
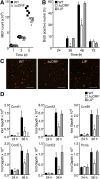
Upstream ORFs (uORFs) are translational control elements found predominantly in transcripts of key regulatory genes. No mammalian genetic model exists to experimentally validate the physiological relevance of uORF-regulated translation initiation. We report that mice deficient for the CCAAT/enhancer-binding protein beta (C/EBPbeta) uORF initiation codon fail to initiate translation of the autoantagonistic LIP (liver inhibitory protein) C/EBPbeta isoform. C/EBPbeta(DeltauORF) mice show hyperactivation of acute-phase response genes, persistent repression of E2F-regulated genes, delayed and blunted S-phase entry of hepatocytes after partial hepatectomy, and impaired osteoclast differentiation. These data and the widespread prevalence of uORFs in mammalian transcriptomes suggest a comprehensive role of uORF-regulated translation in (patho)physiology.
Figures
Similar articles
-
Upstream open reading frames: molecular switches in (patho)physiology.Bioessays. 2010 Oct;32(10):885-93. doi: 10.1002/bies.201000037. Epub 2010 Aug 19. Bioessays. 2010. PMID: 20726009 Free PMC article. Review.
-
Deficiency in mTORC1-controlled C/EBPβ-mRNA translation improves metabolic health in mice.EMBO Rep. 2015 Aug;16(8):1022-36. doi: 10.15252/embr.201439837. Epub 2015 Jun 25. EMBO Rep. 2015. PMID: 26113365 Free PMC article.
-
Inhibition of CCAAT/enhancer-binding protein alpha and beta translation by upstream open reading frames.J Biol Chem. 1998 Apr 17;273(16):9552-60. doi: 10.1074/jbc.273.16.9552. J Biol Chem. 1998. PMID: 9545285
-
A screening strategy for the discovery of drugs that reduce C/EBPβ-LIP translation with potential calorie restriction mimetic properties.Sci Rep. 2017 Feb 15;7:42603. doi: 10.1038/srep42603. Sci Rep. 2017. PMID: 28198412 Free PMC article.
-
Translational Regulation by Upstream Open Reading Frames and Human Diseases.Adv Exp Med Biol. 2019;1157:99-116. doi: 10.1007/978-3-030-19966-1_5. Adv Exp Med Biol. 2019. PMID: 31342439 Review.
Cited by
-
The transcription factor C/EBPβ orchestrates dendritic cell maturation and functionality under homeostatic and malignant conditions.Proc Natl Acad Sci U S A. 2020 Oct 20;117(42):26328-26339. doi: 10.1073/pnas.2008883117. Epub 2020 Oct 5. Proc Natl Acad Sci U S A. 2020. PMID: 33020261 Free PMC article.
-
What Is the Impact of mRNA 5' TL Heterogeneity on Translational Start Site Selection and the Mammalian Cellular Phenotype?Front Genet. 2016 Aug 31;7:156. doi: 10.3389/fgene.2016.00156. eCollection 2016. Front Genet. 2016. PMID: 27630668 Free PMC article. Review.
-
Upstream Open Reading Frames Differentially Regulate Gene-specific Translation in the Integrated Stress Response.J Biol Chem. 2016 Aug 12;291(33):16927-35. doi: 10.1074/jbc.R116.733899. Epub 2016 Jun 29. J Biol Chem. 2016. PMID: 27358398 Free PMC article. Review.
-
C/EBPβ induces B-cell acute lymphoblastic leukemia and cooperates with BLNK mutations.Cancer Sci. 2021 Dec;112(12):4920-4930. doi: 10.1111/cas.15164. Epub 2021 Oct 23. Cancer Sci. 2021. PMID: 34653294 Free PMC article.
-
MicroRNA-191 targets CCAAT/enhanced binding protein β and functions as an oncogenic molecule in human non-small cell lung carcinoma cells.Exp Ther Med. 2019 Aug;18(2):1175-1183. doi: 10.3892/etm.2019.7668. Epub 2019 Jun 13. Exp Ther Med. 2019. PMID: 31316611 Free PMC article.
References
-
- Akira S. IL-6-regulated transcription factors. Int J Biochem Cell Biol. 1997;29:1401–1418. - PubMed
-
- Baer M, Johnson PF. Generation of truncated C/EBPβ isoforms by in vitro proteolysis. J Biol Chem. 2000;275:26582–26590. - PubMed
-
- Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: Unraveling the biology. Trends Biochem Sci. 2004;29:409–417. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
