Population polymorphism of nuclear mitochondrial DNA insertions reveals widespread diploidy associated with loss of heterozygosity in Debaryomyces hansenii
- PMID: 20048048
- PMCID: PMC2837983
- DOI: 10.1128/EC.00263-09
Population polymorphism of nuclear mitochondrial DNA insertions reveals widespread diploidy associated with loss of heterozygosity in Debaryomyces hansenii
Abstract
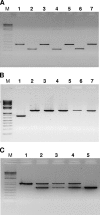
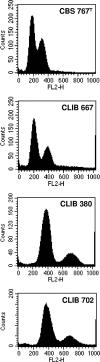
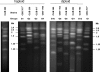
Debaryomyces hansenii, a yeast that participates in the elaboration of foodstuff, displays important genetic diversity. Our recent phylogenetic classification of this species led to the subdivision of the species into three distinct clades. D. hansenii harbors the highest number of nuclear mitochondrial DNA (NUMT) insertions known so far for hemiascomycetous yeasts. Here we assessed the intraspecific variability of the NUMTs in this species by testing their presence/absence first in 28 strains, with 21 loci previously detected in the completely sequenced strain CBS 767(T), and second in a larger panel of 77 strains, with 8 most informative loci. We were able for the first time to structure populations in D. hansenii, although we observed little NUMT insertion variability within the clades. We determined the chronology of the NUMT insertions, which turned out to correlate with the previously defined taxonomy and provided additional evidence that colonization of nuclear genomes by mitochondrial DNA is a dynamic process in yeast. In combination with flow cytometry experiments, the NUMT analysis revealed the existence of both haploid and diploid strains, the latter being heterozygous and resulting from at least four crosses among strains from the various clades. As in the diploid pathogen Candida albicans, to which D. hansenii is phylogenetically related, we observed a differential loss of heterozygosity in the diploid strains, which can explain some of the large genetic diversity found in D. hansenii over the years.
Figures

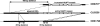
Similar articles
-
Promiscuous DNA in the nuclear genomes of hemiascomycetous yeasts.FEMS Yeast Res. 2008 Sep;8(6):846-57. doi: 10.1111/j.1567-1364.2008.00409.x. Epub 2008 Jul 30. FEMS Yeast Res. 2008. PMID: 18673395
-
Delimitation of the species of the Debaryomyces hansenii complex by intron sequence analysis.Int J Syst Evol Microbiol. 2009 May;59(Pt 5):1242-51. doi: 10.1099/ijs.0.004325-0. Int J Syst Evol Microbiol. 2009. PMID: 19406826
-
DNA typing methods for differentiation of Debaryomyces hansenii strains and other yeasts related to surface ripened cheeses.Int J Food Microbiol. 2001 Sep 19;69(1-2):11-24. doi: 10.1016/s0168-1605(01)00568-2. Int J Food Microbiol. 2001. PMID: 11589549
-
Insertions of mitochondrial DNA into the nucleus-effects and role in cell evolution.Genome. 2020 Aug;63(8):365-374. doi: 10.1139/gen-2019-0151. Epub 2020 May 12. Genome. 2020. PMID: 32396758 Review.
-
Debaryomyces hansenii: an old acquaintance for a fresh start in the era of the green biotechnology.World J Microbiol Biotechnol. 2022 Apr 28;38(6):99. doi: 10.1007/s11274-022-03280-x. World J Microbiol Biotechnol. 2022. PMID: 35482161 Free PMC article. Review.
Cited by
-
Cytogenetic and Sequence Analyses of Mitochondrial DNA Insertions in Nuclear Chromosomes of Maize.G3 (Bethesda). 2015 Sep 1;5(11):2229-39. doi: 10.1534/g3.115.020677. G3 (Bethesda). 2015. PMID: 26333837 Free PMC article.
-
Genetic characterization and construction of an auxotrophic strain of Saccharomyces cerevisiae JP1, a Brazilian industrial yeast strain for bioethanol production.J Ind Microbiol Biotechnol. 2012 Nov;39(11):1673-83. doi: 10.1007/s10295-012-1170-5. Epub 2012 Aug 15. J Ind Microbiol Biotechnol. 2012. PMID: 22892884
-
Insights into the life cycle of yeasts from the CTG clade revealed by the analysis of the Millerozyma (Pichia) farinosa species complex.PLoS One. 2012;7(5):e35842. doi: 10.1371/journal.pone.0035842. Epub 2012 May 4. PLoS One. 2012. PMID: 22574125 Free PMC article.
-
Genome sequence of the highly weak-acid-tolerant Zygosaccharomyces bailii IST302, amenable to genetic manipulations and physiological studies.FEMS Yeast Res. 2017 Jun 1;17(4):fox025. doi: 10.1093/femsyr/fox025. FEMS Yeast Res. 2017. PMID: 28460089 Free PMC article.
-
A practical comparison of the next-generation sequencing platform and assemblers using yeast genome.Life Sci Alliance. 2023 Feb 6;6(4):e202201744. doi: 10.26508/lsa.202201744. Print 2023 Apr. Life Sci Alliance. 2023. PMID: 36746534 Free PMC article.
References
-
- Baroiller C., Schmidt J. L. 1990. Contribution à l'étude de l'origine des levures du fromage de Camembert. Lait 70:67–84
-
- Behura S. K. 2007. Analysis of nuclear copies of mitochondrial sequences in honeybee (Apis mellifera) genome. Mol. Biol. Evol. 24:1492–1505 - PubMed
-
- Bensasson D., Feldman M. W., Petrov D. A. 2003. Rates of DNA duplication and mitochondrial DNA insertion in the human genome. J. Mol. Evol. 57:343–354 - PubMed
-
- Corredor M., Davila A. M., Casaregola S., Gaillardin C. 2003. Chromosomal polymorphism in the yeast species Debaryomyces hansenii. Antonie Van Leeuwenhoek 84:81–88 - PubMed
-
- Corredor M., Davila A. M., Gaillardin C., Casaregola S. 2000. DNA probes specific for the yeast species Debaryomyces hansenii: useful tools for rapid identification. FEMS Microbiol. Lett. 193:171–177 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

