Lactobacillus reuteri 2'-deoxyribosyltransferase, a novel biocatalyst for tailoring of nucleosides
- PMID: 20048065
- PMCID: PMC2832402
- DOI: 10.1128/AEM.01685-09
Lactobacillus reuteri 2'-deoxyribosyltransferase, a novel biocatalyst for tailoring of nucleosides
Abstract
A novel type II nucleoside 2'-deoxyribosyltransferase from Lactobacillus reuteri (LrNDT) has been cloned and overexpressed in Escherichia coli. The recombinant LrNDT has been structural and functionally characterized. Sedimentation equilibrium analysis revealed a homohexameric molecule of 114 kDa. Circular dichroism studies have showed a secondary structure containing 55% alpha-helix, 10% beta-strand, 16% beta-sheet, and 19% random coil. LrNDT was thermostable with a melting temperature (T(m)) of 64 degrees C determined by fluorescence, circular dichroism, and differential scanning calorimetric studies. The enzyme showed high activity in a broad pH range (4.6 to 7.9) and was also very stable between pH 4 and 7.9. The optimal temperature for activity was 40 degrees C. The recombinant LrNDT was able to synthesize natural and nonnatural nucleoside analogues, improving activities described in the literature, and remarkably, exhibited unexpected new arabinosyltransferase activity, which had not been described so far in this kind of enzyme. Furthermore, synthesis of new arabinonucleosides and 2'-fluorodeoxyribonucleosides was carried out.
Figures

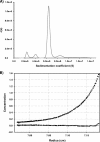

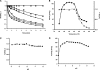

References
-
- Anand, R., P. A. Kaminski, and S. E. Ealick. 2004. Structures of purine 2′-deoxyribosyltransferase, substrate complexes, and the ribosylated enzyme intermediate at 2.0 Å resolution. Biochemistry 43:2384-2393. - PubMed
-
- Armstrong, S. R., W. J. Cook, S. A. Short, and S. E. Ealick. 1996. Crystal structures of nucleoside 2′-deoxyribosyltransferase in native and ligand bound forms reveals architecture of the active site. Structure 4:97. - PubMed
-
- Becker, J., and M. Brendel. 1996. Rapid purification and characterization of two distinct N-deoxyribosyltransferases of Lactobacillus leichmannii. Biol. Chem. Hoppe-Seyler 377:357-362. - PubMed
-
- Bennett, E. M., C. Li, P. W. Allan, W. B. Parker, and S. E. Ealick. 2003. Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase. J. Biol. Chem. 278:47110-47118. - PubMed
-
- Betbeder, D., and L. K. Hutchinson. 1990. The enzymatic synthesis of imidazole deoxynucleosides: 1-β-D-2′-deoxyribofuranosyl-5-aminoimidazole-4-carboxamide and 1-β-D-2′-deoxyribfuranosylbenzimidazoles. Nucleosides Nucleotides Nucleic Acids 9:569-577.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

