Proteoliposomes harboring alkaline phosphatase and nucleotide pyrophosphatase as matrix vesicle biomimetics
- PMID: 20048161
- PMCID: PMC2844207
- DOI: 10.1074/jbc.M109.079830
Proteoliposomes harboring alkaline phosphatase and nucleotide pyrophosphatase as matrix vesicle biomimetics
Abstract
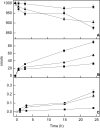
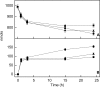
We have established a proteoliposome system as an osteoblast-derived matrix vesicle (MV) biomimetic to facilitate the study of the interplay of tissue-nonspecific alkaline phosphatase (TNAP) and NPP1 (nucleotide pyrophosphatase/phosphodiesterase-1) during catalysis of biomineralization substrates. First, we studied the incorporation of TNAP into liposomes of various lipid compositions (i.e. in pure dipalmitoyl phosphatidylcholine (DPPC), DPPC/dipalmitoyl phosphatidylserine (9:1 and 8:2), and DPPC/dioctadecyl-dimethylammonium bromide (9:1 and 8:2) mixtures. TNAP reconstitution proved virtually complete in DPPC liposomes. Next, proteoliposomes containing either recombinant TNAP, recombinant NPP1, or both together were reconstituted in DPPC, and the hydrolysis of ATP, ADP, AMP, pyridoxal-5'-phosphate (PLP), p-nitrophenyl phosphate, p-nitrophenylthymidine 5'-monophosphate, and PP(i) by these proteoliposomes was studied at physiological pH. p-Nitrophenylthymidine 5'-monophosphate and PLP were exclusively hydrolyzed by NPP1-containing and TNAP-containing proteoliposomes, respectively. In contrast, ATP, ADP, AMP, PLP, p-nitrophenyl phosphate, and PP(i) were hydrolyzed by TNAP-, NPP1-, and TNAP plus NPP1-containing proteoliposomes. NPP1 plus TNAP additively hydrolyzed ATP, but TNAP appeared more active in AMP formation than NPP1. Hydrolysis of PP(i) by TNAP-, and TNAP plus NPP1-containing proteoliposomes occurred with catalytic efficiencies and mild cooperativity, effects comparable with those manifested by murine osteoblast-derived MVs. The reconstitution of TNAP and NPP1 into proteoliposome membranes generates a phospholipid microenvironment that allows the kinetic study of phosphosubstrate catabolism in a manner that recapitulates the native MV microenvironment.
Figures


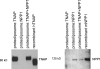

Similar articles
-
Proteoliposomes as matrix vesicles' biomimetics to study the initiation of skeletal mineralization.Braz J Med Biol Res. 2010 Mar;43(3):234-41. doi: 10.1590/s0100-879x2010007500008. Braz J Med Biol Res. 2010. PMID: 20401430 Free PMC article. Review.
-
Effects of pH on the production of phosphate and pyrophosphate by matrix vesicles' biomimetics.Calcif Tissue Int. 2013 Sep;93(3):222-32. doi: 10.1007/s00223-013-9745-3. Epub 2013 May 31. Calcif Tissue Int. 2013. PMID: 23942722 Free PMC article.
-
NPP1 and TNAP hydrolyze ATP synergistically during biomineralization.Purinergic Signal. 2023 Jun;19(2):353-366. doi: 10.1007/s11302-022-09882-2. Epub 2022 Jul 23. Purinergic Signal. 2023. PMID: 35870033 Free PMC article.
-
Kinetic analysis of substrate utilization by native and TNAP-, NPP1-, or PHOSPHO1-deficient matrix vesicles.J Bone Miner Res. 2010 Apr;25(4):716-23. doi: 10.1359/jbmr.091023. J Bone Miner Res. 2010. PMID: 19874193 Free PMC article.
-
The role of phosphatases in the initiation of skeletal mineralization.Calcif Tissue Int. 2013 Oct;93(4):299-306. doi: 10.1007/s00223-012-9672-8. Epub 2012 Nov 27. Calcif Tissue Int. 2013. PMID: 23183786 Free PMC article. Review.
Cited by
-
Proteoliposomes as matrix vesicles' biomimetics to study the initiation of skeletal mineralization.Braz J Med Biol Res. 2010 Mar;43(3):234-41. doi: 10.1590/s0100-879x2010007500008. Braz J Med Biol Res. 2010. PMID: 20401430 Free PMC article. Review.
-
Liposomal systems as carriers for bioactive compounds.Biophys Rev. 2015 Dec;7(4):391-397. doi: 10.1007/s12551-015-0180-8. Epub 2015 Oct 10. Biophys Rev. 2015. PMID: 28510100 Free PMC article. Review.
-
The two sides of a lipid-protein story.Biophys Rev. 2016 Jun;8(2):179-191. doi: 10.1007/s12551-016-0199-5. Epub 2016 Apr 30. Biophys Rev. 2016. PMID: 28510056 Free PMC article. Review.
-
Extracellular pyrophosphate metabolism and calcification in vascular smooth muscle.Am J Physiol Heart Circ Physiol. 2011 Jul;301(1):H61-8. doi: 10.1152/ajpheart.01020.2010. Epub 2011 Apr 13. Am J Physiol Heart Circ Physiol. 2011. PMID: 21490328 Free PMC article.
-
Matrix Vesicles: Role in Bone Mineralization and Potential Use as Therapeutics.Pharmaceuticals (Basel). 2021 Mar 24;14(4):289. doi: 10.3390/ph14040289. Pharmaceuticals (Basel). 2021. PMID: 33805145 Free PMC article. Review.
References
-
- Boskey A. (2006) in Dynamics of Bone and Cartilage Metabolism (Seibel M. J., Robins S. P., Biezikian J. P. eds) pp. 201–212, Academic Press, Inc., New York
-
- Johnson K. A., Hessle L., Vaingankar S., Wennberg C., Mauro S., Narisawa S., Goding J. W., Sano K., Millan J. L., Terkeltaub R. (2000) Am. J. Physiol. Regul. Integr. Comp. Physiol. 279, R1365–R1377 - PubMed
-
- Bernard G. W. (1978) Clin. Orthop. 135, 218–225 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

