Saccharomyces cerevisiae Esc2p interacts with Sir2p through a small ubiquitin-like modifier (SUMO)-binding motif and regulates transcriptionally silent chromatin in a locus-dependent manner
- PMID: 20048165
- PMCID: PMC2844200
- DOI: 10.1074/jbc.M109.016360
Saccharomyces cerevisiae Esc2p interacts with Sir2p through a small ubiquitin-like modifier (SUMO)-binding motif and regulates transcriptionally silent chromatin in a locus-dependent manner
Abstract
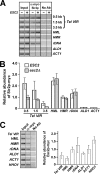
Saccharomyces cerevisiae Esc2p is a member of a conserved family of proteins that contain small ubiquitin-like modifier (SUMO)-like domains. It has been implicated in transcriptional silencing and shown to interact with the silencing protein Sir2p in a two-hybrid analysis. However, little is known about how Esc2p regulates the structure of silent chromatin. We demonstrate here that ESC2 differentially regulates silent chromatin at telomeric, rDNA, and HM loci. Specifically, ESC2 is required for efficient telomeric silencing and Sir2p association with telomeric silent chromatin and for silencing and maintenance of silent chromatin structure at rDNA. On the other hand, ESC2 negatively regulates silencing at HML and HMR and destabilizes HML silent chromatin without affecting Sir2p association with chromatin. We present evidence that Esc2p is associated with both transcriptionally silent and active loci in the genome, and the abundance of Esc2p is not correlated with the chromatin state at a particular locus. Using affinity pull-down analyses, we show that Esc2p and Sir2p interact in vivo, and recombinant Esc2p and Sir2p interact directly. Moreover, we dissect Esc2p and identify a putative SUMO-binding motif that is necessary and sufficient for interacting with Sir2p and SUMO and is required for the function of Esc2p in transcriptional silencing.
Figures


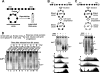



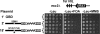
References
-
- Rusche L. N., Kirchmaier A. L., Rine J. (2003) Annu. Rev. Biochem. 72, 481–516 - PubMed
-
- Braunstein M., Rose A. B., Holmes S. G., Allis C. D., Broach J. R. (1993) Genes Dev. 7, 592–604 - PubMed
-
- Moazed D. (2001) Curr. Opin. Cell Biol. 13, 232–238 - PubMed
-
- Suka N., Suka Y., Carmen A.A., Wu J., Grunstein M. (2001) Mol. Cell. 8, 473–479 - PubMed
-
- Shou W., Seol J. H., Shevchenko A., Baskerville C., Moazed D., Chen Z. W., Jang J., Shevchenko A., Charbonneau H., Deshaies R. J. (1999) Cell 97, 233–244 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

