Merging mixture components for cell population identification in flow cytometry
- PMID: 20049161
- PMCID: PMC2798116
- DOI: 10.1155/2009/247646
Merging mixture components for cell population identification in flow cytometry
Abstract
We present a framework for the identification of cell subpopulations in flow cytometry data based on merging mixture components using the flowClust methodology. We show that the cluster merging algorithm under our framework improves model fit and provides a better estimate of the number of distinct cell subpopulations than either Gaussian mixture models or flowClust, especially for complicated flow cytometry data distributions. Our framework allows the automated selection of the number of distinct cell subpopulations and we are able to identify cases where the algorithm fails, thus making it suitable for application in a high throughput FCM analysis pipeline. Furthermore, we demonstrate a method for summarizing complex merged cell subpopulations in a simple manner that integrates with the existing flowClust framework and enables downstream data analysis. We demonstrate the performance of our framework on simulated and real FCM data. The software is available in the flowMerge package through the Bioconductor project.
Figures

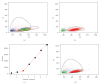

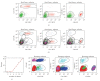
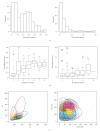



References
-
- Gratama JW, Kraan J, Keeney M, Granger V, Barnett D. Reduction of variation in T-cell subset enumeration among 55 laboratories using single-platform, three or four-color flow cytometry based on CD45 and SSC-based gating of lymphocytes. Clinical Cytometry. 2002;50(2):92–101. - PubMed
-
- Satoh C, Dan K, Yamashita T, Jo R, Tamura H, Ogata K. Flow cytometric parameters with little interexaminer variability for diagnosing low-grade myelodysplastic syndromes. Leukemia Research. 2008;32(5):699–707. - PubMed
-
- Van Blerk M, Bernier M, Bossuyt X, et al. National external quality assessment scheme for lymphocyte immunophenotyping in Belgium. Clinical Chemistry and Laboratory Medicine. 2003;41(3):323–330. - PubMed
-
- Achuthanandam R, Quinn J, Capocasale RJ, Bugelski PJ, Hrebien L, Kam M. Sequential univariate gating approach to study the effects of erythropoietin in murine bone marrow. Cytometry Part A. 2008;73(8):702–714. - PubMed
-
- Boedigheimer MJ, Ferbas J. Mixture modeling approach to flow cytometry data. Cytometry Part A. 2008;73(5):421–429. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

