Predicting the antigenic structure of the pandemic (H1N1) 2009 influenza virus hemagglutinin
- PMID: 20049332
- PMCID: PMC2797400
- DOI: 10.1371/journal.pone.0008553
Predicting the antigenic structure of the pandemic (H1N1) 2009 influenza virus hemagglutinin
Abstract
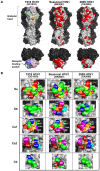
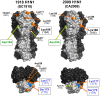
The pandemic influenza virus (2009 H1N1) was recently introduced into the human population. The hemagglutinin (HA) gene of 2009 H1N1 is derived from "classical swine H1N1" virus, which likely shares a common ancestor with the human H1N1 virus that caused the pandemic in 1918, whose descendant viruses are still circulating in the human population with highly altered antigenicity of HA. However, information on the structural basis to compare the HA antigenicity among 2009 H1N1, the 1918 pandemic, and seasonal human H1N1 viruses has been lacking. By homology modeling of the HA structure, here we show that HAs of 2009 H1N1 and the 1918 pandemic virus share a significant number of amino acid residues in known antigenic sites, suggesting the existence of common epitopes for neutralizing antibodies cross-reactive to both HAs. It was noted that the early human H1N1 viruses isolated in the 1930s-1940s still harbored some of the original epitopes that are also found in 2009 H1N1. Interestingly, while 2009 H1N1 HA lacks the multiple N-glycosylations that have been found to be associated with an antigenic change of the human H1N1 virus during the early epidemic of this virus, 2009 H1N1 HA still retains unique three-codon motifs, some of which became N-glycosylation sites via a single nucleotide mutation in the human H1N1 virus. We thus hypothesize that the 2009 H1N1 HA antigenic sites involving the conserved amino acids will soon be targeted by antibody-mediated selection pressure in humans. Indeed, amino acid substitutions predicted here are occurring in the recent 2009 H1N1 variants. The present study suggests that antibodies elicited by natural infection with the 1918 pandemic or its early descendant viruses play a role in specific immunity against 2009 H1N1, and provides an insight into future likely antigenic changes in the evolutionary process of 2009 H1N1 in the human population.
Conflict of interest statement
Figures


Similar articles
-
Multi-task learning sparse group lasso: a method for quantifying antigenicity of influenza A(H1N1) virus using mutations and variations in glycosylation of Hemagglutinin.BMC Bioinformatics. 2020 May 11;21(1):182. doi: 10.1186/s12859-020-3527-5. BMC Bioinformatics. 2020. PMID: 32393178 Free PMC article.
-
Glycosylations in the globular head of the hemagglutinin protein modulate the virulence and antigenic properties of the H1N1 influenza viruses.Sci Transl Med. 2013 May 29;5(187):187ra70. doi: 10.1126/scitranslmed.3005996. Sci Transl Med. 2013. PMID: 23720581 Free PMC article.
-
Minor changes in the hemagglutinin of influenza A(H1N1)2009 virus alter its antigenic properties.PLoS One. 2011;6(10):e25848. doi: 10.1371/journal.pone.0025848. Epub 2011 Oct 11. PLoS One. 2011. PMID: 22022458 Free PMC article.
-
N-linked glycosylation in the hemagglutinin of influenza A viruses.Yonsei Med J. 2012 Sep;53(5):886-93. doi: 10.3349/ymj.2012.53.5.886. Yonsei Med J. 2012. PMID: 22869469 Free PMC article. Review.
-
[Advances in the structure and function of pandemic A/H1N1/2009 influenza virus HA protein].Bing Du Xue Bao. 2012 Jun;28(4):444-52. Bing Du Xue Bao. 2012. PMID: 22978172 Review. Chinese.
Cited by
-
N-linked glycosylation of the hemagglutinin protein influences virulence and antigenicity of the 1918 pandemic and seasonal H1N1 influenza A viruses.J Virol. 2013 Aug;87(15):8756-66. doi: 10.1128/JVI.00593-13. Epub 2013 Jun 5. J Virol. 2013. PMID: 23740978 Free PMC article.
-
Evolutionary, genetic, structural characterization and its functional implications for the influenza A (H1N1) infection outbreak in India from 2009 to 2017.Sci Rep. 2019 Oct 11;9(1):14690. doi: 10.1038/s41598-019-51097-w. Sci Rep. 2019. PMID: 31604969 Free PMC article.
-
Hemagglutinin 222D/G polymorphism facilitates fast intra-host evolution of pandemic (H1N1) 2009 influenza A viruses.PLoS One. 2014 Aug 27;9(8):e104233. doi: 10.1371/journal.pone.0104233. eCollection 2014. PLoS One. 2014. PMID: 25162520 Free PMC article.
-
The Phylodynamics of Seasonal Influenza A/H1N1pdm Virus in China Between 2009 and 2019.Front Microbiol. 2020 Apr 28;11:735. doi: 10.3389/fmicb.2020.00735. eCollection 2020. Front Microbiol. 2020. PMID: 32457705 Free PMC article.
-
Infection Dynamics of Swine Influenza Virus in a Danish Pig Herd Reveals Recurrent Infections with Different Variants of the H1N2 Swine Influenza A Virus Subtype.Viruses. 2020 Sep 10;12(9):1013. doi: 10.3390/v12091013. Viruses. 2020. PMID: 32927910 Free PMC article.
References
-
- Reid AH, Taubenberger JK. The origin of the 1918 pandemic influenza virus: a continuing enigma. J Gen Virol. 2003;84:2285–2292. - PubMed
-
- Vincent AL, Lager KM, Ma W, Lekcharoensuk P, Gramer MR, et al. Evaluation of hemagglutinin subtype 1 swine influenza viruses from the United States. Vet Microbiol. 2006;118:212–222. - PubMed
-
- Sugita S, Yoshioka Y, Itamura S, Kanegae Y, Oguchi K, et al. Molecular evolution of hemagglutinin genes of H1N1 swine and human influenza A viruses. J Mol Evol. 1991;32:16–23. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

