QPCR: a tool for analysis of mitochondrial and nuclear DNA damage in ecotoxicology
- PMID: 20049526
- PMCID: PMC2844971
- DOI: 10.1007/s10646-009-0457-4
QPCR: a tool for analysis of mitochondrial and nuclear DNA damage in ecotoxicology
Abstract
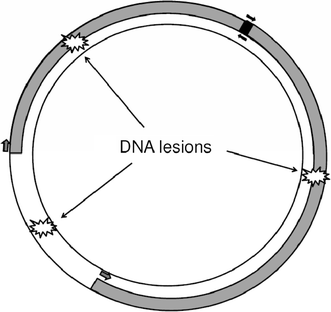
The quantitative PCR (QPCR) assay for DNA damage and repair has been used extensively in laboratory species. More recently, it has been adapted to ecological settings. The purpose of this article is to provide a detailed methodological guide that will facilitate its adaptation to additional species, highlight its potential for ecotoxicological and biomonitoring work, and critically review the strengths and limitations of this assay. Major strengths of the assay include very low (nanogram to picogram) amounts of input DNA; direct comparison of damage and repair in the nuclear and mitochondrial genomes, and different parts of the nuclear genome; detection of a wide range of types of DNA damage; very good reproducibility and quantification; applicability to properly preserved frozen samples; simultaneous monitoring of relative mitochondrial genome copy number; and easy adaptation to most species. Potential limitations include the limit of detection (approximately 1 lesion per 10(5) bases); the inability to distinguish different types of DNA damage; and the need to base quantification of damage on a control or reference sample. I suggest that the QPCR assay is particularly powerful for some ecotoxicological studies.
Figures
Similar articles
-
The long amplicon quantitative PCR for DNA damage assay as a sensitive method of assessing DNA damage in the environmental model, Atlantic killifish (Fundulus heteroclitus).Comp Biochem Physiol C Toxicol Pharmacol. 2009 Mar;149(2):182-6. doi: 10.1016/j.cbpc.2008.07.007. Epub 2008 Jul 24. Comp Biochem Physiol C Toxicol Pharmacol. 2009. PMID: 18706522 Free PMC article.
-
The QPCR assay for analysis of mitochondrial DNA damage, repair, and relative copy number.Methods. 2010 Aug;51(4):444-51. doi: 10.1016/j.ymeth.2010.01.033. Epub 2010 Feb 1. Methods. 2010. PMID: 20123023 Free PMC article. Review.
-
Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells.Methods Mol Biol. 2014;1105:419-37. doi: 10.1007/978-1-62703-739-6_31. Methods Mol Biol. 2014. PMID: 24623245 Free PMC article.
-
Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells.Methods Mol Biol. 2006;314:183-99. doi: 10.1385/1-59259-973-7:183. Methods Mol Biol. 2006. PMID: 16673882
-
Ecotoxicological applications and significance of the comet assay.Mutagenesis. 2008 May;23(3):207-21. doi: 10.1093/mutage/gen014. Epub 2008 Apr 1. Mutagenesis. 2008. PMID: 18381356 Review.
Cited by
-
Neuroligin-mediated neurodevelopmental defects are induced by mitochondrial dysfunction and prevented by lutein in C. elegans.Nat Commun. 2022 May 12;13(1):2620. doi: 10.1038/s41467-022-29972-4. Nat Commun. 2022. PMID: 35551180 Free PMC article.
-
Mitochondrial DNA Damage Does Not Determine C. elegans Lifespan.Front Genet. 2019 Apr 12;10:311. doi: 10.3389/fgene.2019.00311. eCollection 2019. Front Genet. 2019. PMID: 31031801 Free PMC article.
-
Newly Revised Quantitative PCR-Based Assay for Mitochondrial and Nuclear DNA Damage.Curr Protoc Toxicol. 2018 May;76(1):e50. doi: 10.1002/cptx.50. Epub 2018 May 18. Curr Protoc Toxicol. 2018. PMID: 30040241 Free PMC article.
-
Developmental toxicity and DNA damage from exposure to parking lot runoff retention pond samples in the Japanese medaka (Oryzias latipes).Mar Environ Res. 2014 Aug;99:117-24. doi: 10.1016/j.marenvres.2014.04.007. Epub 2014 Apr 26. Mar Environ Res. 2014. PMID: 24816191 Free PMC article.
-
qRT-PCR evaluation of the transcriptional response of zebra mussel to heavy metals.BMC Genomics. 2015 May 6;16(1):354. doi: 10.1186/s12864-015-1567-4. BMC Genomics. 2015. PMID: 25943386 Free PMC article.
References
-
- Anson RM, et al. Mitochondrial endogenous oxidative damage has been overestimated. FASEB J. 2000;14:355–360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

