Heterochromatin protein 1alpha: a hallmark of cell proliferation relevant to clinical oncology
- PMID: 20049717
- PMCID: PMC3378125
- DOI: 10.1002/emmm.200900022
Heterochromatin protein 1alpha: a hallmark of cell proliferation relevant to clinical oncology
Erratum in
- EMBO Mol Med. 2009 Jul;1(4):249
Abstract
Mammalian cells contain three closely related heterochromatin protein 1 (HP1) isoforms, HP1alpha, beta and gamma, which, by analogy to their unique counterpart in Schizosaccharomyces pombe, have been implicated in gene silencing, genome stability and chromosome segregation. However, the individual importance of each isoform during normal cell cycle and disease has remained an unresolved issue. Here, we reveal that HP1alpha shows a proliferation-dependent regulation, which neither HP1beta nor gamma display. During transient cell cycle exit, the HP1alpha mRNA and protein levels diminish. Transient depletion of HP1alpha, but not HP1beta or gamma, in tumoural and primary human cells leads to defects in chromosome segregation. Notably, analysis of an annotated collection of samples derived from carcinomas reveals an overexpression of HP1alpha mRNA and protein, which correlates with clinical data and disease outcome. Our results unveil a specific expression pattern for the HP1alpha isoform, suggesting a unique function related to cell division and tumour growth. The overexpression of HP1alpha constitutes a new example of a potential epigenetic contribution to tumourigenesis that is of clinical interest for cancer prognosis.
Figures

Total protein levels of HP1α, β and γ are detected by Western blot in asynchronously proliferating (As.) and quiescent (G0) WI38 lung fibroblasts, MCF7 breast cancer cells and BJ primary foreskin fibroblasts. Fibroblasts are arrested in quiescence by serum starvation and MCF7 cells by anti-estrogen treatment. Increasing amounts (x) of total cell extracts are loaded and β-actin serves as a loading control. CAF-1 p60 (Polo et al, 2004) and Cyclin A are used as markers for cell proliferation.
Flow cytometry analysis of the cell cycle distribution of the cells shown in A.
HP1α mRNA levels in proliferating (As.) and quiescent (G0) BJ foreskin fibroblasts, as determined by quantitative RT-PCR. Levels are normalized to the reference gene ribosomal protein P0-like protein (RPLPO) (de Cremoux et al, 2004) and levels in proliferating cells are set to 100%. CAF-1 p60 and CAF-1 p150 levels are shown for comparison. The error bar represents data from three independent experiments.

Scheme of the experimental procedure applied to obtain total, soluble and chromatin-bound cell extracts as used in B.
HP1α, β and γ protein levels are analysed by Western blotting in soluble and chromatin-bound nuclear extracts from the breast cancer cell line Hs578T (T) and the non-tumoural mammary cell line Hs578Bst (Bst), which are derived from the same patient (Hackett et al, 1977). Cyclin A and CAF-1 p60 are shown for comparison. Increasing amounts (x) of cell extracts are loaded; β-actin serves as a loading control.
Relative quantification of total HP1α protein levels in tumoural (T) and non-tumoural (Bst) mammary cells, compared to the amounts of CAF-1 p60 (Polo et al, 2004). Increasing amounts (x) of total cell extracts are loaded; β-actin serves as a loading control.
Flow cytometry analysis of tumoural (T) and non-tumoural (Bst) mammary cells in order to assess polyploidy. Tumoural (T) and non-tumoural (Bst) cells contain 25 and 13% of cells in S-phase, respectively.
Relative HP1α mRNA levels in tumoural (T) and non-tumoural (Bst) mammary cells, as determined by quantitative RT-PCR. Levels are normalized to the reference gene ribosomal protein P0-like protein (RPLPO) (de Cremoux et al, 2004) and non-tumoural cells are set to 100%. CAF-1 p150 is shown for comparison. Error bars represent data from two independent experiments.

Western blot analysis of total cell extracts from HeLa cells 72 hours after transfection with siRNA sequences targeting HP1α (named siHP1α.3), HP1β, HP1γ or control siRNA (siGFP). Increasing amounts (x) of total cell extracts are loaded and β-actin is used as a loading control.
Flow cytometry analysis of the cell cycle distribution of cells shown in A.
Typical mitotic figures after transfection with siRNA sequences targeting HP1α, HP1β, HP1γ or mock siRNA (siGFP).
Percentage of aberrant mitotic figures (lagging chromosomes, misalignments, chromosome bridges) in asynchronous HeLa cells 72 h after transfection with siRNA sequences targeting HP1α, HP1β, HP1γ or mock siRNA (siGFP). The error bars represent data from three independent, blind counted, experiments.
DAPI staining reveals micronuclei formation 72 h after transfection with an siRNA sequence targeting HP1α.
Quantification of the percentage of micronucleated cells 72 h after transfection with siRNA sequences targeting HP1α, HP1β, HP1γ or control siRNA (siGFP). Error bars represent data from three independent experiments of which two were blind counted.
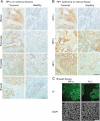
Frozen human tissue arrays containing sections from tumoural and healthy origin are stained for HP1α by immunohistochemistry and counterstained with hematoxylin.
Frozen uterus tissue sections were stained for HP1α, β and γ by immunohistochemistry and counterstained with hematoxylin. Tissue arrays were processed identically for the three antibodies.
Two adjacent sections from a frozen medullary breast carcinoma were stained for HP1α and epithelial pan-cytokeratin marker KL-1, by IF. DNA is revealed with DAPI. Areas of HP1α overexpression correspond to regions of epithelial tumour cells. Scale bars: 100 µm.
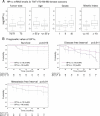
Box plots representing logarithmic HP1α mRNA expression levels, according to the indicated clinico-pathological factors, in 86 breast cancer samples with a >10 year patient follow-up. Boxes represent the 25–75th percentile; brackets: range; black line: median; black dots: outliers.
Univariate Kaplan–Meier curves of the survival, the occurrence of metastasis and the disease-free interval (interval before the occurrence of local recurrence, regional lymph node recurrence, controlateral breast cancer or metastasis) in patients expressing high (>10) or low (≤10) levels of HP1α. The number of patients at risk at each time point is indicated below the graphs.
Similar articles
-
Mammalian HP1 Isoforms Have Specific Roles in Heterochromatin Structure and Organization.Cell Rep. 2017 Nov 21;21(8):2048-2057. doi: 10.1016/j.celrep.2017.10.092. Cell Rep. 2017. PMID: 29166597
-
Beyond the histone tale: HP1α deregulation in breast cancer epigenetics.Cancer Biol Ther. 2015;16(2):189-200. doi: 10.1080/15384047.2014.1001277. Cancer Biol Ther. 2015. PMID: 25588111 Free PMC article. Review.
-
An HP1 isoform-specific feedback mechanism regulates Suv39h1 activity under stress conditions.Epigenetics. 2017 Feb;12(2):166-175. doi: 10.1080/15592294.2016.1278096. Epub 2017 Jan 6. Epigenetics. 2017. PMID: 28059589 Free PMC article.
-
HP1β is a biomarker for breast cancer prognosis and PARP inhibitor therapy.PLoS One. 2015 Mar 13;10(3):e0121207. doi: 10.1371/journal.pone.0121207. eCollection 2015. PLoS One. 2015. PMID: 25769025 Free PMC article.
-
Function of heterochromatin protein 1 during DNA repair.Protoplasma. 2017 May;254(3):1233-1240. doi: 10.1007/s00709-017-1090-3. Epub 2017 Feb 24. Protoplasma. 2017. PMID: 28236007 Review.
Cited by
-
Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer.Nat Rev Cancer. 2012 Oct;12(10):709-20. doi: 10.1038/nrc3344. Epub 2012 Sep 6. Nat Rev Cancer. 2012. PMID: 22952011
-
HP1 promotes tumor suppressor BRCA1 functions during the DNA damage response.Nucleic Acids Res. 2013 Jun;41(11):5784-98. doi: 10.1093/nar/gkt231. Epub 2013 Apr 15. Nucleic Acids Res. 2013. PMID: 23589625 Free PMC article.
-
Heterochromatin Protein 1: A Multiplayer in Cancer Progression.Cancers (Basel). 2022 Feb 1;14(3):763. doi: 10.3390/cancers14030763. Cancers (Basel). 2022. PMID: 35159030 Free PMC article. Review.
-
N-terminal phosphorylation of HP1{alpha} promotes its chromatin binding.Mol Cell Biol. 2011 Mar;31(6):1186-200. doi: 10.1128/MCB.01012-10. Epub 2011 Jan 18. Mol Cell Biol. 2011. PMID: 21245376 Free PMC article.
-
HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair.J Cell Biol. 2011 Apr 4;193(1):81-95. doi: 10.1083/jcb.201101030. J Cell Biol. 2011. PMID: 21464229 Free PMC article.
References
-
- Andersson A, Ritz C, Lindgren D, Eden P, Lassen C, Heldrup J, Olofsson T, Rade J, Fontes M, Porwit-Macdonald A, et al. Microarray-based classification of a consecutive series of 121 childhood acute leukemias: prediction of leukemic and genetic subtype as well as of minimal residual disease status. Leukemia. 2007;21:1198–1203. - PubMed
-
- Auth T, Kunkel E, Grummt F. Interaction between HP1alpha and replication proteins in mammalian cells. Exp Cell Res. 2006;28:28. - PubMed
-
- Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. - PubMed
-
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

