Enzymatic and nonenzymatic functions of viral RNA-dependent RNA polymerases within oligomeric arrays
- PMID: 20051491
- PMCID: PMC2811667
- DOI: 10.1261/rna.1955410
Enzymatic and nonenzymatic functions of viral RNA-dependent RNA polymerases within oligomeric arrays
Abstract
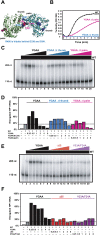
Few antivirals are effective against positive-strand RNA viruses, primarily because the high error rate during replication of these viruses leads to the rapid development of drug resistance. One of the favored current targets for the development of antiviral compounds is the active site of viral RNA-dependent RNA polymerases. However, like many subcellular processes, replication of the genomes of all positive-strand RNA viruses occurs in highly oligomeric complexes on the cytosolic surfaces of the intracellular membranes of infected host cells. In this study, catalytically inactive polymerases were shown to participate productively in functional oligomer formation and catalysis, as assayed by RNA template elongation. Direct protein transduction to introduce either active or inactive polymerases into cells infected with mutant virus confirmed the structural role for polymerase molecules during infection. Therefore, we suggest that targeting the active sites of polymerase molecules is not likely to be the best antiviral strategy, as inactivated polymerases do not inhibit replication of other viruses in the same cell and can, in fact, be useful in RNA replication complexes. On the other hand, polymerases that could not participate in functional RNA replication complexes were those that contained mutations in the amino terminus, leading to altered contacts in the folded polymerase and mutations in a known polymerase-polymerase interaction in the two-dimensional protein lattice. Thus, the functional nature of multimeric arrays of RNA-dependent RNA polymerase supplies a novel target for antiviral compounds and provides a new appreciation for enzymatic catalysis on membranous surfaces within cells.
Figures




Similar articles
-
Structure-function relationships underlying the replication fidelity of viral RNA-dependent RNA polymerases.J Virol. 2015 Jan;89(1):275-86. doi: 10.1128/JVI.01574-14. Epub 2014 Oct 15. J Virol. 2015. PMID: 25320316 Free PMC article.
-
Interstitial contacts in an RNA-dependent RNA polymerase lattice.J Mol Biol. 2011 Sep 30;412(4):737-50. doi: 10.1016/j.jmb.2011.07.053. Epub 2011 Aug 3. J Mol Biol. 2011. PMID: 21839092 Free PMC article.
-
Non-nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase.J Virol. 2020 Oct 14;94(21):e00794-20. doi: 10.1128/JVI.00794-20. Print 2020 Oct 14. J Virol. 2020. PMID: 32796069 Free PMC article.
-
Picornaviral polymerase structure, function, and fidelity modulation.Virus Res. 2017 Apr 15;234:4-20. doi: 10.1016/j.virusres.2017.01.026. Epub 2017 Feb 2. Virus Res. 2017. PMID: 28163093 Free PMC article. Review.
-
The antiviral activity and mechanism of action of (S)-[3-hydroxy-2-(phosphonomethoxy)propyl] (HPMP) nucleosides.Antiviral Res. 2012 Nov;96(2):169-80. doi: 10.1016/j.antiviral.2012.08.010. Epub 2012 Sep 6. Antiviral Res. 2012. PMID: 22960154 Review.
Cited by
-
Quasispecies Nature of RNA Viruses: Lessons from the Past.Vaccines (Basel). 2023 Jan 30;11(2):308. doi: 10.3390/vaccines11020308. Vaccines (Basel). 2023. PMID: 36851186 Free PMC article. Review.
-
Hepatitis E virus RNA-dependent RNA polymerase is involved in RNA replication and infectious particle production.Hepatology. 2022 Jan;75(1):170-181. doi: 10.1002/hep.32100. Epub 2021 Dec 8. Hepatology. 2022. PMID: 34387882 Free PMC article.
-
Negative charge and membrane-tethered viral 3B cooperate to recruit viral RNA dependent RNA polymerase 3D pol.Sci Rep. 2017 Dec 11;7(1):17309. doi: 10.1038/s41598-017-17621-6. Sci Rep. 2017. PMID: 29230036 Free PMC article.
-
Quantifying the roles of space and stochasticity in computer simulations for cell biology and cellular biochemistry.Mol Biol Cell. 2021 Jan 15;32(2):186-210. doi: 10.1091/mbc.E20-08-0530. Epub 2020 Nov 25. Mol Biol Cell. 2021. PMID: 33237849 Free PMC article.
-
RNA-Dependent RNA Polymerases of Picornaviruses: From the Structure to Regulatory Mechanisms.Viruses. 2015 Aug 6;7(8):4438-60. doi: 10.3390/v7082829. Viruses. 2015. PMID: 26258787 Free PMC article. Review.
References
-
- Beckman MT, Kirkegaard K. Site size of cooperative single-stranded RNA binding by poliovirus RNA-dependent RNA polymerase. J Biol Chem. 1998;273:6724–6730. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
