Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries
- PMID: 20051556
- PMCID: PMC2813481
- DOI: 10.1101/gr.095273.109
Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries
Abstract
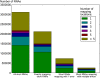
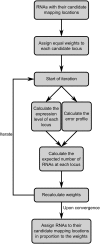
MicroRNAs (miRNAs) are short (20-23 nt) RNAs that are sequence-specific mediators of transcriptional and post-transcriptional regulation of gene expression. Modern high-throughput technologies enable deep sequencing of such RNA species on an unprecedented scale. We find that the analysis of small RNA deep-sequencing libraries can be affected by cross-mapping, in which RNA sequences originating from one locus are inadvertently mapped to another. Similar to cross-hybridization on microarrays, cross-mapping is prevalent among miRNAs, as they tend to occur in families, are similar or derived from repeat or structural RNAs, or are post-transcriptionally modified. Here, we develop a strategy to correct for cross-mapping, and apply it to the analysis of RNA editing in mature miRNAs. In contrast to previous reports, our analysis suggests that RNA editing in mature miRNAs is rare in animals.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
