Molecular cytogenetics of the california condor: evolutionary and conservation implications
- PMID: 20051671
- PMCID: PMC2842169
- DOI: 10.1159/000272458
Molecular cytogenetics of the california condor: evolutionary and conservation implications
Abstract
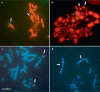
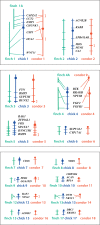
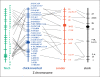
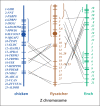
Evolutionary cytogenetic comparisons involved 5 species of birds (California condor, chicken, zebra finch, collared flycatcher and black stork) belonging to divergent taxonomic orders. Seventy-four clones from a condor BAC library containing 80 genes were mapped to condor chromosomes using FISH, and 15 clones containing 16 genes were mapped to the stork Z chromosome. Maps for chicken and finch were derived from genome sequence databases, and that for flycatcher from the published literature. Gene content and gene order were highly conserved when individual condor, chicken, and zebra finch autosomes were compared, confirming that these species largely retain karyotypes close to the ancestral condition for neognathous birds. However, several differences were noted: zebra finch chromosomes 1 and 1A are homologous to condor and chicken chromosomes 1, the CHUNK1 gene appears to have transposed on condor chromosome 1, condor chromosomes 4 and 9 and zebra finch chromosomes 4 and 4A are homologous to chicken chromosome arms 4q and 4p, and novel inversions on chromosomes 4, 12 and 13 were found. Condor and stork Z chromosome gene orders are collinear and differentiated by a series of inversions/transpositions when compared to chicken, zebra finch, or flycatcher; phylogenetic analyses suggest independent rearrangement along the chicken, finch, and flycatcher lineages.
2009 S. Karger AG, Basel.
Figures

References
-
- Belterman RHR, de Boer LEM. A miscellaneous collection of bird karyotypes. Genetica. 1990;83:17–29.
-
- Consortium ICGS. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. - PubMed
-
- de Oliveira EH, Habermann FA, Lacerda O, Sbalqueiro IJ, Wienberg J, Muller S. Chromosome reshuffling in birds of prey: the karyotype of the world's largest eagle (Harpy eagle, Harpia harpyja) compared to that of the chicken (Gallus gallus) Chromosoma. 2005;114:338–343. - PubMed
-
- Fillon V, Vignoles M, Crooijmans RP, Groenen MA, Zoorob R, Vignal A. FISH mapping of 57 BAC clones reveals strong conservation of synteny between Galliformes and Anseriformes. Anim Genet. 2007;38:303–307. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

