DNA resection in eukaryotes: deciding how to fix the break
- PMID: 20051983
- PMCID: PMC2850169
- DOI: 10.1038/nsmb.1710
DNA resection in eukaryotes: deciding how to fix the break
Abstract
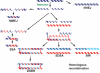
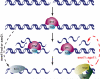
DNA double-strand breaks are repaired by different mechanisms, including homologous recombination and nonhomologous end-joining. DNA-end resection, the first step in recombination, is a key step that contributes to the choice of DSB repair. Resection, an evolutionarily conserved process that generates single-stranded DNA, is linked to checkpoint activation and is critical for survival. Failure to regulate and execute this process results in defective recombination and can contribute to human disease. Here I review recent findings on the mechanisms of resection in eukaryotes, from yeast to vertebrates, provide insights into the regulatory strategies that control it, and highlight the consequences of both its impairment and its deregulation.
Figures

References
-
- Aguilera A, Gómez-González B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. - PubMed
-
- Lieber MR. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2008;283:1–5. - PubMed
-
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

