Differential decay of parent-of-origin-specific genomic sharing in cystic fibrosis-affected sib pairs maps a paternally imprinted locus to 7q34
- PMID: 20051989
- PMCID: PMC2987319
- DOI: 10.1038/ejhg.2009.229
Differential decay of parent-of-origin-specific genomic sharing in cystic fibrosis-affected sib pairs maps a paternally imprinted locus to 7q34
Abstract
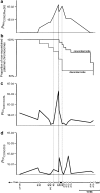
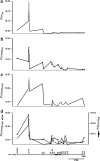
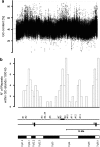
Cystic fibrosis (CF) is a monogenic disease characterized by a high variability of disease severity and outcome that points to the role of environmental factors and modulating genes that shape the course of this multiorgan disease. We genotyped families of cystic fibrosis sib pairs homozygous for F508del-CFTR who represent extreme clinical phenotypes at informative microsatellite markers spanning a 38 Mb region between CFTR and 7qtel. Recombination events on both parental chromosomes were compared between siblings with concordant clinical phenotypes and siblings with discordant clinical phenotypes. Monitoring parent-of-origin-specific decay of genomic sharing delineated a 2.9-Mb segment on 7q34 in which excess of recombination on paternal chromosomes in discordant pairs was observed compared with phenotypically concordant sibs. This 2.9-Mb core candidate region was enriched in imprinting-related elements such as predicted CCCTC-binding factor consensus sites and CpG islands dense in repetitive elements. Moreover, allele frequencies at a microsatellite marker within the core candidate region differed significantly comparing mildly and severely affected cystic fibrosis sib pairs. The identification of this paternally imprinted locus on 7q34 as a modulator of cystic fibrosis disease severity shows that imprinted elements can be identified by straightforward fine mapping of break points in sib pairs with informative contrasting phenotypes.
Figures

References
-
- Reinhart B, Caillet JR. Genomic imprinting: cis-acting sequences and regional control. Int Rev Cytol. 2005;243:173–213. - PubMed
-
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. - PubMed
-
- Knapp M, Strauch K. Affected-sib-pair test for linkage based on constraints for identical-by-descent distributions corresponding to disease models with imprinting. Genet Epidemiol. 2005;26:273–285. - PubMed
-
- Wu CC, Shete S, Amos CI. Linkage analysis of affected sib pairs allowing for parent-of-origin effects. Ann Hum Genet. 2005;69:113–126. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

