Ultradeep sequencing of a human ultraconserved region reveals somatic and constitutional genomic instability
- PMID: 20052272
- PMCID: PMC2794366
- DOI: 10.1371/journal.pbio.1000275
Ultradeep sequencing of a human ultraconserved region reveals somatic and constitutional genomic instability
Abstract
Early detection of cancer-associated genomic instability is crucial, particularly in tumour types in which this instability represents the essential underlying mechanism of tumourigenesis. Currently used methods require the presence of already established neoplastic cells because they only detect clonal mutations. In principle, parallel sequencing of single DNA filaments could reveal the early phases of tumour initiation by detecting low-frequency mutations, provided an adequate depth of coverage and an effective control of the experimental error. We applied ultradeep sequencing to estimate the genomic instability of individuals with hereditary non-polyposis colorectal cancer (HNPCC). To overcome the experimental error, we used an ultraconserved region (UCR) of the human genome as an internal control. By comparing the mutability outside and inside the UCR, we observed a tendency of the ultraconserved element to accumulate significantly fewer mutations than the flanking segments in both neoplastic and nonneoplastic HNPCC samples. No difference between the two regions was detectable in cells from healthy donors, indicating that all three HNPCC samples have mutation rates higher than the healthy genome. This is the first, to our knowledge, direct evidence of an intrinsic genomic instability of individuals with heterozygous mutations in mismatch repair genes, and constitutes the proof of principle for the development of a more sensitive molecular assay of genomic instability.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
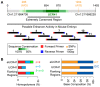
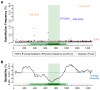
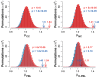

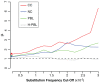
References
-
- Peltomaki P, de la Chapelle A. Mutations predisposing to hereditary nonpolyposis colorectal cancer. Adv Cancer Res. 1997;71:93–119. - PubMed
-
- Loeb L. A. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51:3075–3079. - PubMed
-
- Lynch H. T, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. - PubMed
-
- Aaltonen L. A, Peltomaki P, Mecklin J. P, Jarvinen H, Jass J. R, et al. Replication errors in benign and malignant tumors from hereditary nonpolyposis colorectal cancer patients. Cancer Res. 1994;54:1645–1648. - PubMed
-
- Soreide K. Molecular testing for microsatellite instability and DNA mismatch repair defects in hereditary and sporadic colorectal cancers–ready for prime time? Tumour Biol. 2007;28:290–300. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

