Genetic encoding of non-natural amino acids in Drosophila melanogaster Schneider 2 cells
- PMID: 20052681
- PMCID: PMC2866270
- DOI: 10.1002/pro.322
Genetic encoding of non-natural amino acids in Drosophila melanogaster Schneider 2 cells
Abstract
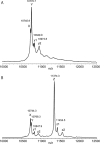
Insect cells are useful for the high-yield production of recombinant proteins including chemokines and membrane proteins. In this study, we developed an insect cell-based system for incorporating non-natural amino acids into proteins at specific sites. Three types of promoter systems were constructed, and their efficiencies were compared for the expression of the prokaryotic amber suppressor tRNA(Tyr) in Drosophila melanogaster Schneider 2 cells. When paired with a variant of Escherichia coli tyrosyl-tRNA synthetase specific for 3-iodo-L-tyrosine, the suppressor tRNA transcribed from the U6 promoter most efficiently incorporated the amino acid into proteins in the cells. The transient and stable introductions of these prokaryotic molecules into the insect cells were then compared in terms of the yield of proteins containing non-natural amino acids, and the "transient" method generated a sevenfold higher yield. By this method, 4-azido-L-phenylalanine was incorporated into human interleukin-8 at a specific site. The yield of the azido-containing IL-8 was 1 microg/1 mL cell culture, and the recombinant protein was successfully labeled with a fluorescent probe by the Staudinger-Bertozzi reaction.
Figures
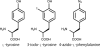
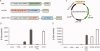


References
-
- Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. - PubMed
-
- Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301:964–967. - PubMed
-
- Xie J, Wang L, Wu N, Brock A, Spraggon G, Schultz PG. The site-specific incorporation of p-iodo-l-phenylalanine into proteins for structure determination. Nat Biotechnol. 2006;22:1297–1301. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

