Refinement of under-determined loops of Human Prion Protein by database-derived distance constraints
- PMID: 20052907
- PMCID: PMC3018887
- DOI: 10.1504/ijdmb.2009.029206
Refinement of under-determined loops of Human Prion Protein by database-derived distance constraints
Abstract
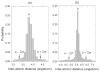
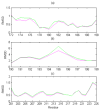
Due to insufficient experimental restraints, a biologically critical loop region in PrP(c) (residues 167-171), which is a potential binding site for Protein X, is under-determined in most mammalian species. Here, we show that by adding information about distance constraints derived from a database of high-resolution protein structures, this under-determined loop as well as other secondary structural elements of the E200K variant of Human Prion Protein (hPrP(c)), a disease-related isoform, can be refined into more realistic structures in the structural ensemble with improved quality and increased accuracy. In particular, the ensemble becomes more compact after the refinement and the percentage of residues in the most favourable region of the Ramachandran diagram is increased to about 90% in the refined structures from the 80% to 85% range in the previously reported structures.
Figures




Similar articles
-
Long-time scale fluctuations of human prion protein determined by restrained MD simulations.Biochemistry. 2011 Nov 29;50(47):10192-4. doi: 10.1021/bi2012756. Epub 2011 Nov 4. Biochemistry. 2011. PMID: 22032174
-
Predicted consequences of site-directed mutagenesis and the impact of species variation on prion protein misfolding through the N-terminal domain.J Mol Model. 2005 Nov;11(6):468-73. doi: 10.1007/s00894-005-0239-8. Epub 2005 Jul 21. J Mol Model. 2005. PMID: 16034619
-
Refinement of NMR-determined protein structures with database derived mean-force potentials.Proteins. 2007 Jul 1;68(1):232-42. doi: 10.1002/prot.21358. Proteins. 2007. PMID: 17387736
-
Computational studies on the prion protein.Curr Top Med Chem. 2013;13(19):2419-31. doi: 10.2174/15680266113136660170. Curr Top Med Chem. 2013. PMID: 24059339 Review.
-
Rare large scale subdomain motions in prion protein can initiate aggregation.J Biomol Struct Dyn. 2006 Jun;23(6):581-90. doi: 10.1080/07391102.2006.10507083. J Biomol Struct Dyn. 2006. PMID: 16615804 Review.
Cited by
-
P.R.E.S.S.--an R-package for exploring residual-level protein structural statistics.J Bioinform Comput Biol. 2012 Jun;10(3):1242007. doi: 10.1142/S0219720012420073. J Bioinform Comput Biol. 2012. PMID: 22809383 Free PMC article.
-
Statistical measures on residue-level protein structural properties.J Struct Funct Genomics. 2011 Jul;12(2):119-36. doi: 10.1007/s10969-011-9104-4. Epub 2011 Mar 31. J Struct Funct Genomics. 2011. PMID: 21452025 Free PMC article.
References
-
- Brown P, Gajdusek DC. The human spongiform encephalopathies: kuru, Creutzfeldt-Jakob disease, and the Gerstmann-Straussler-Scheinker syndrome. Curr Top Microbiol Immunol. 1991;171:1–20. - PubMed
-
- Horwich AL, Weissman JS. Deadly conformations – protein misfolding in prion disease. Cell. 1997;89:499–510. - PubMed
-
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. - PubMed
-
- Stahl N, Baldwin MA, Teplow DB, Hood L, Gibson BW, Burlingame AL, Prusiner SB. Structural studies of the scrapie prion protein using mass spectrometry and amino acid sequencing. Biochemistry. 1993;32:1991–2002. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
