PRTAD: a database for protein residue torsion angle distributions
- PMID: 20052908
- PMCID: PMC3018885
- DOI: 10.1504/ijdmb.2009.029207
PRTAD: a database for protein residue torsion angle distributions
Abstract
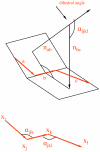
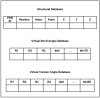
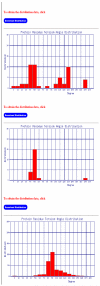
PRTAD is a dedicated database and structural bioinformatics system for protein analysis and modelling. The database is developed to host and analyse the statistical data for protein residue level 'virtual' bond and torsion angles obtained from their distributions in databases of known protein structures such as in the PDB Data Bank. PRTAD is capable of generating, caching, and displaying the statistical distributions of the angles of various types. The collected information can be used to extract geometric restraints or define statistical potentials for protein structure determination. PRTAD is supported with a friendly designed web interface so that users can easily specify the angle types, and retrieve, visualise, or download the distributions of the angles as they desire.
Figures







Similar articles
-
PIDD: database for Protein Inter-atomic Distance Distributions.Nucleic Acids Res. 2007 Jan;35(Database issue):D202-7. doi: 10.1093/nar/gkl802. Epub 2006 Dec 6. Nucleic Acids Res. 2007. PMID: 17151078 Free PMC article.
-
PPInterface: A Comprehensive Dataset of 3D Protein-Protein Interface Structures.J Mol Biol. 2024 Sep 1;436(17):168686. doi: 10.1016/j.jmb.2024.168686. Epub 2024 Jun 25. J Mol Biol. 2024. PMID: 38936693
-
PDB-tools web: A user-friendly interface for the manipulation of PDB files.Proteins. 2021 Mar;89(3):330-335. doi: 10.1002/prot.26018. Epub 2020 Nov 7. Proteins. 2021. PMID: 33111403 Free PMC article.
-
Protein structure databases with new web services for structural biology and biomedical research.Brief Bioinform. 2008 Jul;9(4):276-85. doi: 10.1093/bib/bbn015. Epub 2008 Apr 22. Brief Bioinform. 2008. PMID: 18430752 Review.
-
Recent developments of the chemistry development kit (CDK) - an open-source java library for chemo- and bioinformatics.Curr Pharm Des. 2006;12(17):2111-20. doi: 10.2174/138161206777585274. Curr Pharm Des. 2006. PMID: 16796559 Review.
Cited by
-
P.R.E.S.S.--an R-package for exploring residual-level protein structural statistics.J Bioinform Comput Biol. 2012 Jun;10(3):1242007. doi: 10.1142/S0219720012420073. J Bioinform Comput Biol. 2012. PMID: 22809383 Free PMC article.
-
Statistical measures on residue-level protein structural properties.J Struct Funct Genomics. 2011 Jul;12(2):119-36. doi: 10.1007/s10969-011-9104-4. Epub 2011 Mar 31. J Struct Funct Genomics. 2011. PMID: 21452025 Free PMC article.
References
-
- Creighton TE. Proteins: Structures and Molecular Properties. 2nd Edition Freeman and Company; 1993.
-
- Cui F, Jernigan R, Wu Zj. Refinement of NMR-determined protein structures with database derived distance constraints. J Bioinformatics and Computational Biology. 2005;3:1315–1329. - PubMed
-
- Wu D, Jernigan R, Wu Z. Refinement of NMR-determined protein structures with database derived mean force potentials. Proteins: Struct. Funct. Bioinf. 2007 published online, DOI: 10.1002/prot.21358. - PubMed
-
- Feng Y, Kloczkowski A, Jernigan R. Four-body contact potentials derived from two protein databases to discriminate native structures from decoys. Proteins: Struct. Funct. Bioinf. 2007 published online, DOI: 10.1002/prot.21362. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
