Statistical method on nonrandom clustering with application to somatic mutations in cancer
- PMID: 20053295
- PMCID: PMC2822753
- DOI: 10.1186/1471-2105-11-11
Statistical method on nonrandom clustering with application to somatic mutations in cancer
Abstract
Background: Human cancer is caused by the accumulation of tumor-specific mutations in oncogenes and tumor suppressors that confer a selective growth advantage to cells. As a consequence of genomic instability and high levels of proliferation, many passenger mutations that do not contribute to the cancer phenotype arise alongside mutations that drive oncogenesis. While several approaches have been developed to separate driver mutations from passengers, few approaches can specifically identify activating driver mutations in oncogenes, which are more amenable for pharmacological intervention.
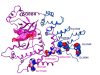
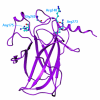
Results: We propose a new statistical method for detecting activating mutations in cancer by identifying nonrandom clusters of amino acid mutations in protein sequences. A probability model is derived using order statistics assuming that the location of amino acid mutations on a protein follows a uniform distribution. Our statistical measure is the differences between pair-wise order statistics, which is equivalent to the size of an amino acid mutation cluster, and the probabilities are derived from exact and approximate distributions of the statistical measure. Using data in the Catalog of Somatic Mutations in Cancer (COSMIC) database, we have demonstrated that our method detects well-known clusters of activating mutations in KRAS, BRAF, PI3K, and beta-catenin. The method can also identify new cancer targets as well as gain-of-function mutations in tumor suppressors.
Conclusions: Our proposed method is useful to discover activating driver mutations in cancer by identifying nonrandom clusters of somatic amino acid mutations in protein sequences.
Figures
Similar articles
-
Utilizing protein structure to identify non-random somatic mutations.BMC Bioinformatics. 2013 Jun 13;14:190. doi: 10.1186/1471-2105-14-190. BMC Bioinformatics. 2013. PMID: 23758891 Free PMC article.
-
Use of signals of positive and negative selection to distinguish cancer genes and passenger genes.Elife. 2021 Jan 11;10:e59629. doi: 10.7554/eLife.59629. Elife. 2021. PMID: 33427197 Free PMC article.
-
Mutational screening of RET, HRAS, KRAS, NRAS, BRAF, AKT1, and CTNNB1 in medullary thyroid carcinoma.Anticancer Res. 2011 Dec;31(12):4179-83. Anticancer Res. 2011. PMID: 22199277
-
[Current topics in mutations in the cancer genome].Nihon Geka Gakkai Zasshi. 2012 Mar;113(2):185-90. Nihon Geka Gakkai Zasshi. 2012. PMID: 22582578 Review. Japanese.
-
Therapeutic strategies for targeting BRAF in human cancer.Rev Recent Clin Trials. 2007 May;2(2):121-34. doi: 10.2174/157488707780599393. Rev Recent Clin Trials. 2007. PMID: 18473997 Review.
Cited by
-
Proteome-Wide Assessment of Clustering of Missense Variants in Neurodevelopmental Disorders Versus Cancer.medRxiv [Preprint]. 2024 Feb 4:2024.02.02.24302238. doi: 10.1101/2024.02.02.24302238. medRxiv. 2024. Update in: Cell Genom. 2025 Apr 9;5(4):100807. doi: 10.1016/j.xgen.2025.100807. PMID: 38352539 Free PMC article. Updated. Preprint.
-
Proteome-wide assessment of differential missense variant clustering in neurodevelopmental disorders and cancer.Cell Genom. 2025 Apr 9;5(4):100807. doi: 10.1016/j.xgen.2025.100807. Epub 2025 Mar 11. Cell Genom. 2025. PMID: 40073865 Free PMC article.
-
PertInInt: An Integrative, Analytical Approach to Rapidly Uncover Cancer Driver Genes with Perturbed Interactions and Functionalities.Cell Syst. 2020 Jul 22;11(1):63-74.e7. doi: 10.1016/j.cels.2020.06.005. Epub 2020 Jul 14. Cell Syst. 2020. PMID: 32711844 Free PMC article.
-
Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes.Nat Genet. 2015 Feb;47(2):106-14. doi: 10.1038/ng.3168. Epub 2014 Dec 15. Nat Genet. 2015. PMID: 25501392 Free PMC article.
-
Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples.Genet Med. 2017 Feb;19(2):192-203. doi: 10.1038/gim.2016.90. Epub 2016 Aug 17. Genet Med. 2017. PMID: 27532257 Free PMC article.
References
-
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, Wendl MC, Lawrence MS, Larson DE, Chen K, Dooling DJ, Sabo A, Hawes AC, Shen H, Jhangiani SN, Lewis LR, Hall O, Zhu Y, Mathew T, Ren Y, Yao J, Scherer SE, Clerc K, Metcalf GA, Ng B, Milosavljevic A, Gonzalez-Garay ML, Osborne JR, Meyer R, Shi X, Tang Y, Koboldt DC, Lin L, Abbott R, Miner TL, Pohl C, Fewell G, Haipek C, Schmidt H, Dunford-Shore BH, Kraja A, Crosby SD, Sawyer CS, Vickery T, Sander S, Robinson J, Winckler W, Baldwin J, Chirieac LR, Dutt A, Fennell T, Hanna M, Johnson BE, Onofrio RC, Thomas RK, Tonon G, Weir BA, Zhao X, Ziaugra L, Zody MC, Giordano T, Orringer MB, Roth JA, Spitz MR, Wistuba II, Ozenberger B, Good PJ, Chang AC, Beer DG, Watson MA, Ladanyi M, Broderick S, Yoshizawa A, Travis WD, Pao W, Province MA, Weinstock GM, Varmus HE, Gabriel SB, Lander ES, Gibbs RA, Meyerson M, Wilson RK. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

