The PIN-FORMED (PIN) protein family of auxin transporters
- PMID: 20053306
- PMCID: PMC2812941
- DOI: 10.1186/gb-2009-10-12-249
The PIN-FORMED (PIN) protein family of auxin transporters
Abstract
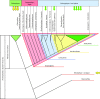
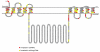
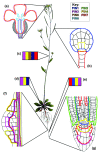
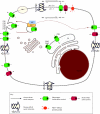
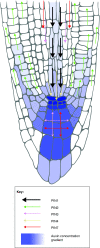
The PIN-FORMED (PIN) proteins are secondary transporters acting in the efflux of the plant signal molecule auxin from cells. They are asymmetrically localized within cells and their polarity determines the directionality of intercellular auxin flow. PIN genes are found exclusively in the genomes of multicellular plants and play an important role in regulating asymmetric auxin distribution in multiple developmental processes, including embryogenesis, organogenesis, tissue differentiation and tropic responses. All PIN proteins have a similar structure with amino- and carboxy-terminal hydrophobic, membrane-spanning domains separated by a central hydrophilic domain. The structure of the hydrophobic domains is well conserved. The hydrophilic domain is more divergent and it determines eight groups within the protein family. The activity of PIN proteins is regulated at multiple levels, including transcription, protein stability, subcellular localization and transport activity. Different endogenous and environmental signals can modulate PIN activity and thus modulate auxin-distribution-dependent development. A large group of PIN proteins, including the most ancient members known from mosses, localize to the endoplasmic reticulum and they regulate the subcellular compartmentalization of auxin and thus auxin metabolism. Further work is needed to establish the physiological importance of this unexpected mode of auxin homeostasis regulation. Furthermore, the evolution of PIN-based transport, PIN protein structure and more detailed biochemical characterization of the transport function are important topics for further studies.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

