Conformational dynamics and structural plasticity play critical roles in the ubiquitin recognition of a UIM domain
- PMID: 20053359
- PMCID: PMC2854419
- DOI: 10.1016/j.jmb.2009.12.052
Conformational dynamics and structural plasticity play critical roles in the ubiquitin recognition of a UIM domain
Abstract
Ubiquitin-interacting motifs (UIMs) are an important class of protein domains that interact with ubiquitin or ubiquitin-like proteins. These approximately 20-residue-long domains are found in a variety of ubiquitin receptor proteins and serve as recognition modules towards intracellular targets, which may be individual ubiquitin subunits or polyubiquitin chains attached to a variety of proteins. Previous structural studies of interactions between UIMs and ubiquitin have shown that UIMs adopt an extended structure of a single alpha-helix, containing a hydrophobic surface with a conserved sequence pattern that interacts with key hydrophobic residues on ubiquitin. In light of this large body of structural studies, details regarding the presence and the roles of structural dynamics and plasticity are surprisingly lacking. In order to better understand the structural basis of ubiquitin-UIM recognition, we have characterized changes in the structure and dynamics of ubiquitin upon binding of a UIM domain from the yeast Vps27 protein. The solution structure of a ubiquitin-UIM fusion protein designed to study these interactions is reported here and found to consist of a well-defined ubiquitin core and a bipartite UIM helix. Moreover, we have studied the plasticity of the docking interface, as well as global changes in ubiquitin due to UIM binding at the picoseconds-to-nanoseconds and microseconds-to-milliseconds protein motions by nuclear magnetic resonance relaxation. Changes in generalized-order parameters of amide groups show a distinct trend towards increased structural rigidity at the UIM-ubiquitin interface relative to values determined in unbound ubiquitin. Analysis of (15)N Carr-Purcell-Meiboom-Gill relaxation dispersion measurements suggests the presence of two types of motions: one directly related to the UIM-binding interface and the other induced to distal parts of the protein. This study demonstrates a case where localized interactions among protein domains have global effects on protein motions at timescales ranging from picoseconds to milliseconds.
(c) 2009 Elsevier Ltd. All rights reserved.
Figures

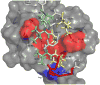
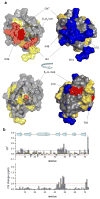
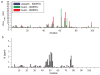

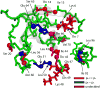
Similar articles
-
Structural and functional characterization of a ubiquitin variant engineered for tight and specific binding to an alpha-helical ubiquitin interacting motif.Protein Sci. 2017 May;26(5):1060-1069. doi: 10.1002/pro.3155. Epub 2017 Mar 24. Protein Sci. 2017. PMID: 28276594 Free PMC article.
-
Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation.EMBO J. 2003 Sep 15;22(18):4597-606. doi: 10.1093/emboj/cdg471. EMBO J. 2003. PMID: 12970172 Free PMC article.
-
A ubiquitin-interacting motif from Hrs binds to and occludes the ubiquitin surface necessary for polyubiquitination in monoubiquitinated proteins.Biochem Biophys Res Commun. 2002 Sep 6;296(5):1222-7. doi: 10.1016/s0006-291x(02)02006-5. Biochem Biophys Res Commun. 2002. PMID: 12207904
-
Solution structure of UIM and interaction of tandem ubiquitin binding domains in STAM1 with ubiquitin.Biochem Biophys Res Commun. 2011 Feb 4;405(1):24-30. doi: 10.1016/j.bbrc.2010.12.103. Epub 2010 Dec 25. Biochem Biophys Res Commun. 2011. PMID: 21187078
-
Ubiquitin--molecular mechanisms for recognition of different structures.Curr Opin Struct Biol. 2010 Jun;20(3):367-76. doi: 10.1016/j.sbi.2010.03.007. Epub 2010 Apr 22. Curr Opin Struct Biol. 2010. PMID: 20456943 Review.
Cited by
-
Differential proteomics study of postharvest Volvariella volvacea during storage at 4 °C.Sci Rep. 2020 Aug 4;10(1):13134. doi: 10.1038/s41598-020-69988-8. Sci Rep. 2020. PMID: 32753745 Free PMC article.
-
Dimeric interactions and complex formation using direct coevolutionary couplings.Sci Rep. 2015 Sep 4;5:13652. doi: 10.1038/srep13652. Sci Rep. 2015. PMID: 26338201 Free PMC article.
-
Ubiquitin Interacting Motifs: Duality Between Structured and Disordered Motifs.Front Mol Biosci. 2021 Jun 28;8:676235. doi: 10.3389/fmolb.2021.676235. eCollection 2021. Front Mol Biosci. 2021. PMID: 34262938 Free PMC article.
-
Bacterial expression and purification of the amyloidogenic peptide PAPf39 for multidimensional NMR spectroscopy.Protein Expr Purif. 2013 Apr;88(2):196-200. doi: 10.1016/j.pep.2013.01.003. Epub 2013 Jan 11. Protein Expr Purif. 2013. PMID: 23314347 Free PMC article.
-
In silico elucidation of the recognition dynamics of ubiquitin.PLoS Comput Biol. 2011 Apr;7(4):e1002035. doi: 10.1371/journal.pcbi.1002035. Epub 2011 Apr 21. PLoS Comput Biol. 2011. PMID: 21533067 Free PMC article.
References
-
- Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–21. - PubMed
-
- Hofmann K, Falquet L. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem Sci. 2001;26:347–50. - PubMed
-
- Young P, Deveraux Q, Beal RE, Pickart CM, Rechsteiner M. Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a. J Biol Chem. 1998;273:5461–7. - PubMed
-
- Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annual Review of Biochemistry. 2003;72:395–447. - PubMed
-
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–6. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

