Desolvation and development of specific hydrophobic core packing during Im7 folding
- PMID: 20053361
- PMCID: PMC2833379
- DOI: 10.1016/j.jmb.2009.12.048
Desolvation and development of specific hydrophobic core packing during Im7 folding
Abstract
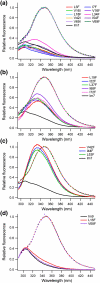
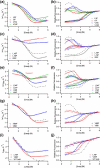
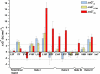
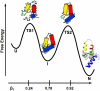
Development of a tightly packed hydrophobic core drives the folding of water-soluble globular proteins and is a key determinant of protein stability. Despite this, there remains much to be learnt about how and when the hydrophobic core becomes desolvated and tightly packed during protein folding. We have used the bacterial immunity protein Im7 to examine the specificity of hydrophobic core packing during folding. This small, four-helix protein has previously been shown to fold via a compact three-helical intermediate state. Here, overpacking substitutions, in which residue side-chain size is increased, were used to examine the specificity and malleability of core packing in the folding intermediate and rate-limiting transition state. In parallel, polar groups were introduced into the Im7 hydrophobic core via Val-->Thr or Phe-->Tyr substitutions and used to determine the solvation status of core residues at different stages of folding. Over 30 Im7 variants were created allowing both series of substitutions to cover all regions of the protein structure. Phi-value analysis demonstrated that the major changes in Im7 core solvation occur prior to the population of the folding intermediate, with key regions involved in docking of the short helix III remaining solvent-exposed until after the rate-limiting transition state has been traversed. In contrast, overpacking core residues revealed that some regions of the native Im7 core are remarkably malleable to increases in side-chain volume. Overpacking residues in other regions of the Im7 core result in substantial (>2.5 kJ mol(-1)) destabilisation of the native structure or even prevents efficient folding to the native state. This study provides new insights into Im7 folding; demonstrating that whilst desolvation occurs early during folding, adoption of a specifically packed core is achieved only at the very last step in the folding mechanism.
Copyright (c) 2010 Elsevier Ltd. All rights reserved.
Figures







Similar articles
-
Dissecting key residues in folding and stability of the bacterial immunity protein 7.Protein Eng Des Sel. 2011 Jun;24(6):517-23. doi: 10.1093/protein/gzr009. Epub 2011 Mar 10. Protein Eng Des Sel. 2011. PMID: 21393384 Free PMC article.
-
Amino acid insertion reveals a necessary three-helical intermediate in the folding pathway of the colicin E7 immunity protein Im7.J Mol Biol. 2009 Oct 2;392(4):1074-86. doi: 10.1016/j.jmb.2009.07.085. Epub 2009 Aug 3. J Mol Biol. 2009. PMID: 19651139 Free PMC article.
-
NMR analysis of the conformational properties of the trapped on-pathway folding intermediate of the bacterial immunity protein Im7.J Mol Biol. 2007 Feb 23;366(3):1001-15. doi: 10.1016/j.jmb.2006.11.012. Epub 2006 Nov 10. J Mol Biol. 2007. PMID: 17188712 Free PMC article.
-
beta-hairpin-forming peptides; models of early stages of protein folding.Biophys Chem. 2010 Sep;151(1-2):1-9. doi: 10.1016/j.bpc.2010.05.001. Epub 2010 May 6. Biophys Chem. 2010. PMID: 20494507 Free PMC article. Review.
-
Folding speeds of helical membrane proteins.Biochem Soc Trans. 2024 Feb 28;52(1):491-501. doi: 10.1042/BST20231315. Biochem Soc Trans. 2024. PMID: 38385525 Free PMC article. Review.
Cited by
-
Dissecting key residues in folding and stability of the bacterial immunity protein 7.Protein Eng Des Sel. 2011 Jun;24(6):517-23. doi: 10.1093/protein/gzr009. Epub 2011 Mar 10. Protein Eng Des Sel. 2011. PMID: 21393384 Free PMC article.
-
Perturbing the folding energy landscape of the bacterial immunity protein Im7 by site-specific N-linked glycosylation.Proc Natl Acad Sci U S A. 2010 Dec 28;107(52):22528-33. doi: 10.1073/pnas.1015356107. Epub 2010 Dec 9. Proc Natl Acad Sci U S A. 2010. PMID: 21148421 Free PMC article.
-
The folding of single domain proteins--have we reached a consensus?Curr Opin Struct Biol. 2011 Feb;21(1):12-24. doi: 10.1016/j.sbi.2010.11.002. Epub 2010 Dec 6. Curr Opin Struct Biol. 2011. PMID: 21144739 Free PMC article. Review.
-
Native contact density and nonnative hydrophobic effects in the folding of bacterial immunity proteins.PLoS Comput Biol. 2015 May 27;11(5):e1004260. doi: 10.1371/journal.pcbi.1004260. eCollection 2015 May. PLoS Comput Biol. 2015. PMID: 26016652 Free PMC article.
-
Energetic frustrations in protein folding at residue resolution: a homologous simulation study of Im9 proteins.PLoS One. 2014 Jan 31;9(1):e87719. doi: 10.1371/journal.pone.0087719. eCollection 2014. PLoS One. 2014. PMID: 24498176 Free PMC article.
References
-
- Kellis J.T., Jr., Nyberg K., Sali D., Fersht A.R. Contribution of hydrophobic interactions to protein stability. Nature. 1988;333:784–786. - PubMed
-
- Baldwin R.L. Energetics of protein folding. J. Mol. Biol. 2007;371:283–301. - PubMed
-
- Ventura S., Serrano L. Designing proteins from the inside out. Proteins. 2004;56:1–10. - PubMed
-
- Matouschek A., Kellis J.T., Serrano L., Fersht A.R. Mapping the transition state and pathway of protein folding by protein engineering. Nature. 1989;340:122–126. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

