The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing
- PMID: 20053671
- PMCID: PMC2838533
- DOI: 10.1093/hmg/ddp585
The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing
Abstract
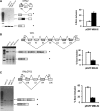
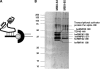
The loss of HBII-52 and related C/D box small nucleolar RNA (snoRNA) expression units have been implicated as a cause for the Prader-Willi syndrome (PWS). We recently found that the C/D box snoRNA HBII-52 changes the alternative splicing of the serotonin receptor 2C pre-mRNA, which is different from the traditional C/D box snoRNA function in non-mRNA methylation. Using bioinformatic predictions and experimental verification, we identified five pre-mRNAs (DPM2, TAF1, RALGPS1, PBRM1 and CRHR1) containing alternative exons that are regulated by MBII-52, the mouse homolog of HBII-52. Analysis of a single member of the MBII-52 cluster of snoRNAs by RNase protection and northern blot analysis shows that the MBII-52 expressing unit generates shorter RNAs that originate from the full-length MBII-52 snoRNA through additional processing steps. These novel RNAs associate with hnRNPs and not with proteins associated with canonical C/D box snoRNAs. Our data indicate that not a traditional C/D box snoRNA MBII-52, but a processed version lacking the snoRNA stem is the predominant MBII-52 RNA missing in PWS. This processed snoRNA functions in alternative splice-site selection. Its substitution could be a therapeutic principle for PWS.
Figures





References
-
- Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. - PubMed
-
- Stamm S., Ben-Ari S., Rafalska I., Tang Y., Zhang Z., Toiber D., Thanaraj T.A., Soreq H. Function of alternative splicing. Gene. 2005;344C:1–20. - PubMed
-
- Watkins N.J., Segault V., Charpentier B., Nottrott S., Fabrizio P., Bachi A., Wilm M., Rosbash M., Branlant C., Luhrmann R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. - PubMed
-
- Matera A.G., Terns R.M., Terns M.P. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

