Numb independently antagonizes Sanpodo membrane targeting and Notch signaling in Drosophila sensory organ precursor cells
- PMID: 20053677
- PMCID: PMC2828966
- DOI: 10.1091/mbc.e09-09-0831
Numb independently antagonizes Sanpodo membrane targeting and Notch signaling in Drosophila sensory organ precursor cells
Abstract
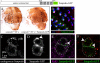
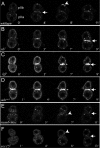
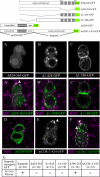
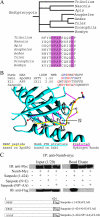
In Drosophila, mitotic neural progenitor cells asymmetrically segregate the cell fate determinant Numb in order to block Notch signaling in only one of the two daughter cells. Sanpodo, a membrane protein required for Notch signaling in asymmetrically dividing cells, is sequestered from the plasma membrane to intracellular vesicles in a Numb-dependent way after neural progenitor cell mitosis. However, the significance of Numb-dependent Sanpodo regulation is unclear. In this study, we conducted a structure-function analysis to identify the determinants of Sanpodo targeting in vivo. We identified an NPAF motif in the amino-terminal cytoplasmic tail of Sanpodo, which is conserved among insect Sanpodo homologues. The Sanpodo NPAF motif is predicted to bind directly to the Numb phosphotyrosine-binding domain and is critical for Numb binding in vitro. Deletion or mutation of the NPAF motif results in accumulation of Sanpodo at the plasma membrane in Numb-positive cells in vivo. Genetic analysis of Sanpodo NPAF mutants shows that Numb-dependent Sanpodo endocytic targeting can be uncoupled from Notch signaling regulation. Our findings demonstrate that Sanpodo contains an evolutionarily conserved motif that has been linked to Numb-dependent regulation in vertebrates and further support the model that Numb regulates Notch signaling independently of Sanpodo membrane trafficking in neural progenitor cells.
Figures
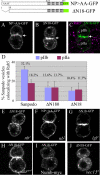
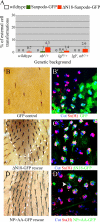
Similar articles
-
The Multitasker Protein: A Look at the Multiple Capabilities of NUMB.Cells. 2023 Jan 15;12(2):333. doi: 10.3390/cells12020333. Cells. 2023. PMID: 36672267 Free PMC article. Review.
-
Numb and alpha-Adaptin regulate Sanpodo endocytosis to specify cell fate in Drosophila external sensory organs.EMBO Rep. 2005 Sep;6(9):836-42. doi: 10.1038/sj.embor.7400500. EMBO Rep. 2005. PMID: 16113648 Free PMC article.
-
Numb inhibits membrane localization of Sanpodo, a four-pass transmembrane protein, to promote asymmetric divisions in Drosophila.Dev Cell. 2003 Aug;5(2):231-43. doi: 10.1016/s1534-5807(03)00226-0. Dev Cell. 2003. PMID: 12919675
-
Regulation of membrane localization of Sanpodo by lethal giant larvae and neuralized in asymmetrically dividing cells of Drosophila sensory organs.Mol Biol Cell. 2005 Aug;16(8):3480-7. doi: 10.1091/mbc.e05-03-0177. Epub 2005 May 18. Mol Biol Cell. 2005. PMID: 15901829 Free PMC article.
-
Endocytosis and control of Notch signaling.Curr Opin Cell Biol. 2012 Aug;24(4):534-40. doi: 10.1016/j.ceb.2012.06.006. Epub 2012 Jul 18. Curr Opin Cell Biol. 2012. PMID: 22818956 Free PMC article. Review.
Cited by
-
Membrane traffic research: challenges for the next decade.Front Cell Dev Biol. 2014 Sep 17;2:52. doi: 10.3389/fcell.2014.00052. eCollection 2014. Front Cell Dev Biol. 2014. PMID: 25364759 Free PMC article. No abstract available.
-
Golgi during development.Cold Spring Harb Perspect Biol. 2011 Sep 1;3(9):a005363. doi: 10.1101/cshperspect.a005363. Cold Spring Harb Perspect Biol. 2011. PMID: 21768608 Free PMC article. Review.
-
The Multitasker Protein: A Look at the Multiple Capabilities of NUMB.Cells. 2023 Jan 15;12(2):333. doi: 10.3390/cells12020333. Cells. 2023. PMID: 36672267 Free PMC article. Review.
-
Cargo recognition in clathrin-mediated endocytosis.Cold Spring Harb Perspect Biol. 2013 Nov 1;5(11):a016790. doi: 10.1101/cshperspect.a016790. Cold Spring Harb Perspect Biol. 2013. PMID: 24186068 Free PMC article. Review.
-
Adaptor proteins involved in polarized sorting.J Cell Biol. 2014 Jan 6;204(1):7-17. doi: 10.1083/jcb.201310021. J Cell Biol. 2014. PMID: 24395635 Free PMC article. Review.
References
-
- Berdnik D., Torok T., Gonzalez-Gaitan M., Knoblich J. A. The endocytic protein alpha-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev. Cell. 2002;3:221–231. - PubMed
-
- Brand A. H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
-
- Calderwood D. A., Fujioka Y., de Pereda J. M., Garcia-Alvarez B., Nakamoto T., Margolis B., McGlade C. J., Liddington R. C., Ginsberg M. H. Integrin beta cytoplasmic domain interactions with phosphotyrosine-binding domains: a structural prototype for diversity in integrin signaling. Proc. Natl. Acad. Sci. USA. 2003;100:2272–2277. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

