The DR1 and DR6 first exons of human herpesvirus 6A are not required for virus replication in culture and are deleted in virus stocks that replicate well in T-cell lines
- PMID: 20053742
- PMCID: PMC2826055
- DOI: 10.1128/JVI.01951-09
The DR1 and DR6 first exons of human herpesvirus 6A are not required for virus replication in culture and are deleted in virus stocks that replicate well in T-cell lines
Abstract
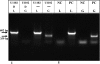
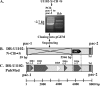
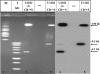
Human herpesvirus 6A (HHV-6A) and HHV-6B are lymphotropic viruses which replicate in cultured activated cord blood mononuclear cells (CBMCs) and in T-cell lines. Viral genomes are composed of 143-kb unique (U) sequences flanked by approximately 8- to 10-kb left and right direct repeats, DR(L) and DR(R). We have recently cloned HHV-6A (U1102) into bacterial artificial chromosome (BAC) vectors, employing DNA replicative intermediates. Surprisingly, HHV-6A BACs and their parental DNAs were found to contain short approximately 2.7-kb DRs. To test whether DR shortening occurred during passaging in CBMCs or in the SupT1 T-cell line, we compared packaged DNAs from various passages. Restriction enzymes, PCR, and sequencing analyses have shown the following. (i) Early (1992) viral preparations from CBMCs contained approximately 8-kb DRs. (ii) Viruses currently propagated in SupT1 cells contained approximately 2.7-kb DRs. (iii) The deletion spans positions 60 to 5545 in DR(L), including genes encoded by DR1 through the first exon of DR6. The pac-2-pac-1 packaging signals, the DR7 open reading frame (ORF), and the DR6 second exon were not deleted. (iv) The DR(R) sequence was similarly shortened by 5.4 kb. (v) The DR1 through DR6 first exon sequences were deleted from the entire HHV-6A BACs, revealing that they were not translocated into other genome locations. (vi) When virus initially cultured in CBMCs was passaged in SupT1 cells no DR shortening occurred. (vii) Viral stocks possessing short DRs replicated efficiently, revealing the plasticity of herpesvirus genomes. We conclude that the DR deletion occurred once, producing virus with advantageous growth "conquering" the population. The DR1 gene and the first DR6 exon are not required for propagation in culture.
Figures




Similar articles
-
Copy Number Heterogeneity, Large Origin Tandem Repeats, and Interspecies Recombination in Human Herpesvirus 6A (HHV-6A) and HHV-6B Reference Strains.J Virol. 2018 Apr 27;92(10):e00135-18. doi: 10.1128/JVI.00135-18. Print 2018 May 15. J Virol. 2018. PMID: 29491155 Free PMC article.
-
Cloning human herpes virus 6A genome into bacterial artificial chromosomes and study of DNA replication intermediates.Proc Natl Acad Sci U S A. 2009 Nov 10;106(45):19138-43. doi: 10.1073/pnas.0908504106. Epub 2009 Oct 26. Proc Natl Acad Sci U S A. 2009. PMID: 19858479 Free PMC article.
-
Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A.J Virol. 1999 Oct;73(10):8040-52. doi: 10.1128/JVI.73.10.8040-8052.1999. J Virol. 1999. PMID: 10482553 Free PMC article.
-
Molecular biology of human herpesviruses 6A and 6B.Infect Agents Dis. 1993 Dec;2(6):343-60. Infect Agents Dis. 1993. PMID: 8012736 Review.
-
Characterization of the lymphotropic amplicons-6 and tamplicon-7 vectors derived from HHV-6 and HHV-7.Curr Gene Ther. 2006 Jun;6(3):399-420. doi: 10.2174/156652306777592036. Curr Gene Ther. 2006. PMID: 16787191 Review.
Cited by
-
Excision of Integrated Human Herpesvirus 6A Genomes Using CRISPR/Cas9 Technology.Microbiol Spectr. 2023 Mar 16;11(2):e0076423. doi: 10.1128/spectrum.00764-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 36926973 Free PMC article.
-
Reactivation of chromosomally integrated human herpesvirus-6 by telomeric circle formation.PLoS Genet. 2013;9(12):e1004033. doi: 10.1371/journal.pgen.1004033. Epub 2013 Dec 19. PLoS Genet. 2013. PMID: 24367281 Free PMC article.
-
Copy Number Heterogeneity, Large Origin Tandem Repeats, and Interspecies Recombination in Human Herpesvirus 6A (HHV-6A) and HHV-6B Reference Strains.J Virol. 2018 Apr 27;92(10):e00135-18. doi: 10.1128/JVI.00135-18. Print 2018 May 15. J Virol. 2018. PMID: 29491155 Free PMC article.
-
Mapping the telomere integrated genome of human herpesvirus 6A and 6B.Virology. 2013 Jul 20;442(1):3-11. doi: 10.1016/j.virol.2013.03.030. Epub 2013 May 4. Virology. 2013. PMID: 23648233 Free PMC article.
-
Human herpesvirus 6 glycoprotein complex formation is required for folding and trafficking of the gH/gL/gQ1/gQ2 complex and its cellular receptor binding.J Virol. 2011 Nov;85(21):11121-30. doi: 10.1128/JVI.05251-11. Epub 2011 Aug 17. J Virol. 2011. PMID: 21849437 Free PMC article.
References
-
- Ablashi, D., H. Agut, Z. Berneman, et al. 1993. Human herpesvirus-6 strain groups: a nomenclature. Arch. Virol. 129:363-366. - PubMed
-
- Achour, A., I. Malet, C. Deback, P. Bonnafous, D. Boutolleau, A. Gautheret-Dejean, and H. Agut. 2009. Length variability of telomeric repeat sequences of human herpesvirus 6 DNA. J. Virol. Methods 159:127-130. - PubMed
-
- Carrigan, D. R., W. R. Drobyski, S. K. Russler, M. A. Tapper, K. K. Knox, and R. C. Ash. 1991. Interstitial pneumonitis associated with human herpesvirus-6 infection after marrow transplantation. Lancet 338:147-149. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

