Bioinformatics challenges for genome-wide association studies
- PMID: 20053841
- PMCID: PMC2820680
- DOI: 10.1093/bioinformatics/btp713
Bioinformatics challenges for genome-wide association studies
Abstract
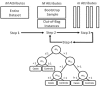
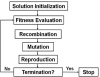
The sequencing of the human genome has made it possible to identify an informative set of >1 million single nucleotide polymorphisms (SNPs) across the genome that can be used to carry out genome-wide association studies (GWASs). The availability of massive amounts of GWAS data has necessitated the development of new biostatistical methods for quality control, imputation and analysis issues including multiple testing. This work has been successful and has enabled the discovery of new associations that have been replicated in multiple studies. However, it is now recognized that most SNPs discovered via GWAS have small effects on disease susceptibility and thus may not be suitable for improving health care through genetic testing. One likely explanation for the mixed results of GWAS is that the current biostatistical analysis paradigm is by design agnostic or unbiased in that it ignores all prior knowledge about disease pathobiology. Further, the linear modeling framework that is employed in GWAS often considers only one SNP at a time thus ignoring their genomic and environmental context. There is now a shift away from the biostatistical approach toward a more holistic approach that recognizes the complexity of the genotype-phenotype relationship that is characterized by significant heterogeneity and gene-gene and gene-environment interaction. We argue here that bioinformatics has an important role to play in addressing the complexity of the underlying genetic basis of common human diseases. The goal of this review is to identify and discuss those GWAS challenges that will require computational methods.
Figures
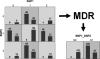
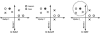
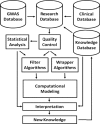
References
-
- Andrew AS, et al. Concordance of multiple analytical approaches demonstrates a complex relationship between DNA repair gene SNPs, smoking and bladder cancer susceptibility. Carcinogenesis. 2006;27:1030–1037. - PubMed
-
- Askland K, et al. Pathways-based analyses of whole-genome association study data in bipolar disorder reveal genes mediating ion channel activity and synaptic neurotransmission. Hum. Genet. 2009;125:63–79. - PubMed

