Flexible recognition of the tRNA G18 methylation target site by TrmH methyltransferase through first binding and induced fit processes
- PMID: 20053984
- PMCID: PMC2838323
- DOI: 10.1074/jbc.M109.065698
Flexible recognition of the tRNA G18 methylation target site by TrmH methyltransferase through first binding and induced fit processes
Abstract
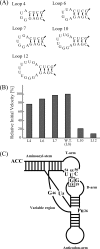
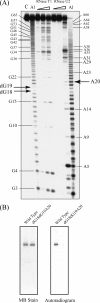
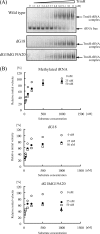
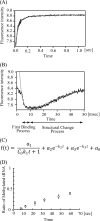
Transfer RNA (Gm18) methyltransferase (TrmH) catalyzes methyl transfer from S-adenosyl-l-methionine to a conserved G18 in tRNA. We investigated the recognition mechanism of Thermus thermophilus TrmH for its guanosine target. Thirteen yeast tRNA(Phe) mutant transcripts were prepared in which the modification site and/or other nucleotides in the D-loop were substituted by dG, inosine, or other nucleotides. We then conducted methyl transfer kinetic studies, gel shift assays, and inhibition experiments using these tRNA variants. Sites of methylation were confirmed with RNA sequencing or primer extension. Although the G18G19 sequence is not essential for methylation by TrmH, disruption of G18G19 severely reduces the efficiency of methyl transfer. There is strict recognition of guanosine by TrmH, in that methylation occurs at the adjacent G19 when the G18 is replaced by dG or adenosine. The fact that TrmH methylates guanosine in D-loops from 4 to 12 nucleotides in length suggests that selection of the position of guanosine within the D-loop is relatively flexible. Our studies also demonstrate that the oxygen 6 atom of the guanine base is a positive determinant for TrmH recognition. The recognition process of TrmH for substrate is inducible and product-inhibited, in that tRNAs containing Gm18 are excluded by TrmH. In contrast, substitution of G18 with dG18 results in the formation of a more stable TrmH-tRNA complex. To address the mechanism, we performed the stopped-flow pre-steady state kinetic analysis. The result clearly showed that the binding of TrmH to tRNA is composed of at least three steps, the first bi-molecular binding and the subsequent two uni-molecular induced-fit processes.
Figures




Similar articles
-
The catalytic domain of topological knot tRNA methyltransferase (TrmH) discriminates between substrate tRNA and nonsubstrate tRNA via an induced-fit process.J Biol Chem. 2013 Aug 30;288(35):25562-25574. doi: 10.1074/jbc.M113.485128. Epub 2013 Jul 18. J Biol Chem. 2013. PMID: 23867454 Free PMC article.
-
Escherichia coli tRNA (Gm18) methyltransferase (TrmH) requires the correct localization of its methylation site (G18) in the D-loop for efficient methylation.J Biochem. 2023 Dec 20;175(1):43-56. doi: 10.1093/jb/mvad076. J Biochem. 2023. PMID: 37844264 Free PMC article.
-
Essentially minimal sequence for substrate recognition by tRNA (guanosine-2')-methyltransferase from Thermus thermophilus HB27.Nucleic Acids Symp Ser. 1997;(37):189-90. Nucleic Acids Symp Ser. 1997. PMID: 9586063
-
Transfer RNA methyltransferases with a SpoU-TrmD (SPOUT) fold and their modified nucleosides in tRNA.Biomolecules. 2017 Feb 28;7(1):23. doi: 10.3390/biom7010023. Biomolecules. 2017. PMID: 28264529 Free PMC article. Review.
-
Enzymatic formation of N2,N2-dimethylguanosine in eukaryotic tRNA: importance of the tRNA architecture.Biochimie. 1995;77(1-2):54-61. doi: 10.1016/0300-9084(96)88104-1. Biochimie. 1995. PMID: 7599276 Review.
Cited by
-
New substrates and determinants for tRNA recognition of RNA methyltransferase DNMT2/TRDMT1.RNA Biol. 2021 Dec;18(12):2531-2545. doi: 10.1080/15476286.2021.1930756. Epub 2021 Jun 10. RNA Biol. 2021. PMID: 34110975 Free PMC article.
-
Unexpected expansion of tRNA substrate recognition by the yeast m1G9 methyltransferase Trm10.RNA. 2013 Aug;19(8):1137-46. doi: 10.1261/rna.039651.113. Epub 2013 Jun 21. RNA. 2013. PMID: 23793893 Free PMC article.
-
RNA Modifications Modulate Activation of Innate Toll-Like Receptors.Genes (Basel). 2019 Jan 29;10(2):92. doi: 10.3390/genes10020092. Genes (Basel). 2019. PMID: 30699960 Free PMC article. Review.
-
A modified dinucleotide motif specifies tRNA recognition by TLR7.RNA. 2014 Sep;20(9):1351-5. doi: 10.1261/rna.044024.113. Epub 2014 Jul 22. RNA. 2014. PMID: 25051971 Free PMC article.
-
Methylated nucleosides in tRNA and tRNA methyltransferases.Front Genet. 2014 May 23;5:144. doi: 10.3389/fgene.2014.00144. eCollection 2014. Front Genet. 2014. PMID: 24904644 Free PMC article. Review.
References
-
- Garcia G., R., Goodenough-Lashhua D. M. (1998) in Modification and Editing of RNA (Grosjean H., Benne R. eds) pp. 555–560, American Society for Microbiology, Washington, D. C.
-
- Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. (1974) Nature 250, 546–551 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

