Stochastic spatio-temporal dynamic model for gene/protein interaction network in early Drosophila development
- PMID: 20054403
- PMCID: PMC2796968
Stochastic spatio-temporal dynamic model for gene/protein interaction network in early Drosophila development
Abstract
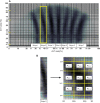
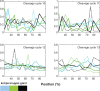
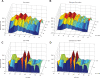
In order to investigate the possible mechanisms for eve stripe formation of Drosophila embryo, a spatio-temporal gene/protein interaction network model is proposed to mimic dynamic behaviors of protein synthesis, protein decay, mRNA decay, protein diffusion, transcription regulations and autoregulation to analyze the interplay of genes and proteins at different compartments in early embryogenesis. In this study, we use the maximum likelihood (ML) method to identify the stochastic 3-D Embryo Space-Time (3-DEST) dynamic model for gene/protein interaction network via 3-D mRNA and protein expression data and then use the Akaike Information Criterion (AIC) to prune the gene/protein interaction network. The identified gene/protein interaction network allows us not only to analyze the dynamic interplay of genes and proteins on the border of eve stripes but also to infer that eve stripes are established and maintained by network motifs built by the cooperation between transcription regulations and diffusion mechanisms in early embryogenesis. Literature reference with the wet experiments of gene mutations provides a clue for validating the identified network. The proposed spatio-temporal dynamic model can be extended to gene/protein network construction of different biological phenotypes, which depend on compartments, e.g. postnatal stem/progenitor cell differentiation.
Keywords: 3-D embryo space-time dynamic model; diffusion mechanism; drosophila embryo; eve stripe formation; gene/protein interaction network; transcription regulation.
Figures


Similar articles
-
Temporal dynamics of pair-rule stripes in living Drosophila embryos.Proc Natl Acad Sci U S A. 2018 Aug 14;115(33):8376-8381. doi: 10.1073/pnas.1810430115. Epub 2018 Jul 30. Proc Natl Acad Sci U S A. 2018. PMID: 30061421 Free PMC article.
-
Mechanism of eve stripe formation.Mech Dev. 1995 Jan;49(1-2):133-58. doi: 10.1016/0925-4773(94)00310-j. Mech Dev. 1995. PMID: 7748785 Review.
-
Quantitative analysis reveals genotype- and domain- specific differences between mRNA and protein expression of segmentation genes in Drosophila.Dev Biol. 2019 Apr 1;448(1):48-58. doi: 10.1016/j.ydbio.2019.01.006. Epub 2019 Jan 7. Dev Biol. 2019. PMID: 30629954 Free PMC article.
-
Transcriptional bursting in Drosophila development: Stochastic dynamics of eve stripe 2 expression.PLoS One. 2017 Apr 24;12(4):e0176228. doi: 10.1371/journal.pone.0176228. eCollection 2017. PLoS One. 2017. PMID: 28437444 Free PMC article.
-
Stripe formation in the early fly embryo: principles, models, and networks.Bioessays. 2009 Nov;31(11):1172-80. doi: 10.1002/bies.200900096. Bioessays. 2009. PMID: 19795410 Review.
Cited by
-
On the Interplay between the Evolvability and Network Robustness in an Evolutionary Biological Network: A Systems Biology Approach.Evol Bioinform Online. 2011;7:201-33. doi: 10.4137/EBO.S8123. Epub 2011 Nov 1. Evol Bioinform Online. 2011. PMID: 22084563 Free PMC article.
-
Integrated cellular and gene interaction modelling of pattern formation.Int J Comput Biol Drug Des. 2011;4(4):361-72. doi: 10.1504/IJCBDD.2011.044444. Epub 2011 Dec 24. Int J Comput Biol Drug Des. 2011. PMID: 22199036 Free PMC article.
References
-
- Driever W, Nusslein-Volhard C. A gradient of bicoid protein in Drosophila embryos. Cell. 1988;54:83–93. - PubMed
-
- Eldar A, Dorfman R, Weiss D, Ashe H, Shilo BZ, Barkai N. Robustness of the BMP morphogen gradient in Drosophila embryonic patterning. Nature. 2002;419:304–8. - PubMed
-
- Gilbert SF.Developmental biology Sinauer Associates, Inc; Publishers, Sunderland, Mass. 2006
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
