A review of computational methods in materials science: examples from shock-wave and polymer physics
- PMID: 20054467
- PMCID: PMC2801990
- DOI: 10.3390/ijms10125135
A review of computational methods in materials science: examples from shock-wave and polymer physics
Abstract
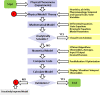
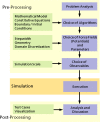
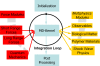
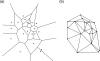
This review discusses several computational methods used on different length and time scales for the simulation of material behavior. First, the importance of physical modeling and its relation to computer simulation on multiscales is discussed. Then, computational methods used on different scales are shortly reviewed, before we focus on the molecular dynamics (MD) method. Here we survey in a tutorial-like fashion some key issues including several MD optimization techniques. Thereafter, computational examples for the capabilities of numerical simulations in materials research are discussed. We focus on recent results of shock wave simulations of a solid which are based on two different modeling approaches and we discuss their respective assets and drawbacks with a view to their application on multiscales. Then, the prospects of computer simulations on the molecular length scale using coarse-grained MD methods are covered by means of examples pertaining to complex topological polymer structures including star-polymers, biomacromolecules such as polyelectrolytes and polymers with intrinsic stiffness. This review ends by highlighting new emerging interdisciplinary applications of computational methods in the field of medical engineering where the application of concepts of polymer physics and of shock waves to biological systems holds a lot of promise for improving medical applications such as extracorporeal shock wave lithotripsy or tumor treatment.
Keywords: biopolymers; computational physics; dendrimers; lithotripsy; modeling and simulation; molecular dynamics; multiscale methods; polymers; shock waves.
Figures




























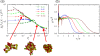
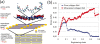

Similar articles
-
How to predict diffusion of medium-sized molecules in polymer matrices. From atomistic to coarse grain simulations.J Mol Model. 2010 Dec;16(12):1845-51. doi: 10.1007/s00894-010-0687-7. Epub 2010 Mar 12. J Mol Model. 2010. PMID: 20224911
-
Coarse-graining in polymer simulation: from the atomistic to the mesoscopic scale and back.Chemphyschem. 2002 Sep 16;3(9):755-69. doi: 10.1002/1439-7641(20020916)3:9<754::aid-cphc754>3.0.co;2-u. Chemphyschem. 2002. PMID: 12436902 Review.
-
An update on the use of molecular modeling in dendrimers design for biomedical applications: are we using its full potential?Expert Opin Drug Discov. 2020 Sep;15(9):1015-1024. doi: 10.1080/17460441.2020.1769597. Epub 2020 May 26. Expert Opin Drug Discov. 2020. PMID: 32452244 Review.
-
A Review of Multiscale Computational Methods in Polymeric Materials.Polymers (Basel). 2017 Jan 9;9(1):16. doi: 10.3390/polym9010016. Polymers (Basel). 2017. PMID: 30970697 Free PMC article. Review.
-
An FDTD-based computer simulation platform for shock wave propagation in electrohydraulic lithotripsy.Comput Methods Programs Biomed. 2013 Jun;110(3):389-98. doi: 10.1016/j.cmpb.2012.11.011. Epub 2012 Dec 20. Comput Methods Programs Biomed. 2013. PMID: 23261077
Cited by
-
Thermal Conductivity Enhancement of Doped Magnesium Hydroxide for Medium-Temperature Heat Storage: A Molecular Dynamics Approach and Experimental Validation.Int J Mol Sci. 2024 Oct 17;25(20):11139. doi: 10.3390/ijms252011139. Int J Mol Sci. 2024. PMID: 39456921 Free PMC article.
-
Solvation Free Energy of Self-Assembled Complexes: Using Molecular Dynamics to Understand the Separation of ssDNA-Wrapped Single-Walled Carbon Nanotubes.J Phys Chem C Nanomater Interfaces. 2020;124:10.1021/acs.jpcc.0c00983. doi: 10.1021/acs.jpcc.0c00983. J Phys Chem C Nanomater Interfaces. 2020. PMID: 34136061 Free PMC article.
-
Gaussian-Accelerated Molecular Dynamics with the Weighted Ensemble Method: A Hybrid Method Improves Thermodynamic and Kinetic Sampling.J Chem Theory Comput. 2021 Dec 14;17(12):7938-7951. doi: 10.1021/acs.jctc.1c00770. Epub 2021 Nov 30. J Chem Theory Comput. 2021. PMID: 34844409 Free PMC article.
-
Research on metallic glasses at the atomic scale: a systematic review.SN Appl Sci. 2022;4(10):281. doi: 10.1007/s42452-022-05170-1. Epub 2022 Sep 30. SN Appl Sci. 2022. PMID: 36196063 Free PMC article. Review.
-
A Review on the Modeling of the Elastic Modulus and Yield Stress of Polymers and Polymer Nanocomposites: Effect of Temperature, Loading Rate and Porosity.Polymers (Basel). 2022 Jan 18;14(3):360. doi: 10.3390/polym14030360. Polymers (Basel). 2022. PMID: 35160350 Free PMC article. Review.
References
-
- Phillips R. Crystals, Defects and Microstructures—Modeling Across Scales. Cambridge University Press; Cambridge, UK: 2001.
-
- Yip S, editor. Handbook of Materials Modeling. Springer; Berlin, Germany: 2005.
-
- Steinhauser MO. Computational Multiscale Modeling of Solids and Fluids—Theory and Applications. Springer; Berlin/Heidelberg/New York, Germany/USA: 2008.
-
- Hockney R, Eastwood J. Computer Simulation Using Particles. McGraw-Hill; New York, NY, USA: 1981.
-
- Ciccotti G, Frenkel G, McDonald I. Simulation of Liquids and Solids. North-Holland; Amsterdam, The Netherlands: 1987.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

