The solution structure of the Mg2+ form of soybean calmodulin isoform 4 reveals unique features of plant calmodulins in resting cells
- PMID: 20054830
- PMCID: PMC2866273
- DOI: 10.1002/pro.325
The solution structure of the Mg2+ form of soybean calmodulin isoform 4 reveals unique features of plant calmodulins in resting cells
Abstract
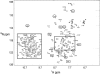
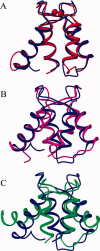
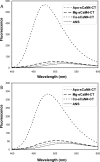
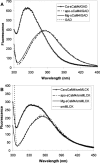
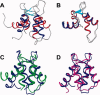
Soybean calmodulin isoform 4 (sCaM4) is a plant calcium-binding protein, regulating cellular responses to the second messenger Ca(2+). We have found that the metal ion free (apo-) form of sCaM4 possesses a half unfolded structure, with the N-terminal domain unfolded and the C-terminal domain folded. This result was unexpected as the apo-forms of both soybean calmodulin isoform 1 (sCaM1) and mammalian CaM (mCaM) are fully folded. Because of the fact that free Mg(2+) ions are always present at high concentrations in cells (0.5-2 mM), we suggest that Mg(2+) should be bound to sCaM4 in nonactivated cells. CD studies revealed that in the presence of Mg(2+) the initially unfolded N-terminal domain of sCaM4 folds into an alpha-helix-rich structure, similar to the Ca(2+) form. We have used the NMR backbone residual dipolar coupling restraints (1)D(NH), (1)D(C alpha H alpha), and (1)D(C'C alpha) to determine the solution structure of the N-terminal domain of Mg(2+)-sCaM4 (Mg(2+)-sCaM4-NT). Compared with the known structure of Ca(2+)-sCaM4, the structure of the Mg(2+)-sCaM4-NT does not fully open the hydrophobic pocket, which was further confirmed by the use of the fluorescent probe ANS. Tryptophan fluorescence experiments were used to study the interactions between Mg(2+)-sCaM4 and CaM-binding peptides derived from smooth muscle myosin light chain kinase and plant glutamate decarboxylase. These results suggest that Mg(2+)-sCaM4 does not bind to Ca(2+)-CaM target peptides and therefore is functionally similar to apo-mCaM. The Mg(2+)- and apo-structures of the sCaM4-NT provide unique insights into the structure and function of some plant calmodulins in resting cells.
Figures

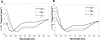
References
-
- Yamniuk AP, Vogel HJ. Calmodulin's flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol Biotechnol. 2004;27:33–57. - PubMed
-
- Barbato G, Ikura M, Kay LE, Pastor RW, Bax A. Backbone dynamics of calmodulin studied by N-15 relaxation using inverse detected 2-dimensional NMR-spectroscopy—the central helix is flexible. Biochemistry. 1992;31:5269–5278. - PubMed
-
- Henikoff S, Greene EA, Pietrokovski S, Bork P, Attwood TK, Hood L. Gene families: the taxonomy of protein paralogs and chimeras. Science. 1997;278:609–614. - PubMed
-
- Gifford JL, Walsh MP, Vogel HJ. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem J. 2007;405:199–221. - PubMed
-
- Wilson MA, Brunger AT. The 1.0 angstrom crystal structure of Ca2+-bound calmodulan analysis of disorder and implications for functionally relevant plasticity. J Mol Biol. 2000;301:1237–1256. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

