Functional motifs in biochemical reaction networks
- PMID: 20055671
- PMCID: PMC3773234
- DOI: 10.1146/annurev.physchem.012809.103457
Functional motifs in biochemical reaction networks
Abstract
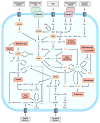
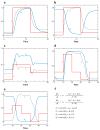
The signal-response characteristics of a living cell are determined by complex networks of interacting genes, proteins, and metabolites. Understanding how cells respond to specific challenges, how these responses are contravened in diseased cells, and how to intervene pharmacologically in the decision-making processes of cells requires an accurate theory of the information-processing capabilities of macromolecular regulatory networks. Adopting an engineer's approach to control systems, we ask whether realistic cellular control networks can be decomposed into simple regulatory motifs that carry out specific functions in a cell. We show that such functional motifs exist and review the experimental evidence that they control cellular responses as expected.
Figures





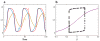


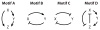

References
-
- Bray D. Protein molecules as computational elements in living cells. Nature. 1995;376:307–12. - PubMed
-
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–61. - PubMed
-
- Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. Boca Raton, FL: Chapman & Hall/CRC; 2007.
-
- Thomas R, D’Ari R. Biological Feedback. Boca Raton, FL: CRC; 1990.
-
- Wolf DM, Arkin AP. Motifs, modules and games in bacteria. Curr Opin Microbiol. 2003;6:125–34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

